Optimization of Medium Components and Genes Expression Involved in IAA Biosynthesis by Serratia plymuthica UBCF_13
DOI:
https://doi.org/10.48048/tis.2024.7852Keywords:
HPLC, Indole-3-acetic acid, Medium optimization, Plant growth promoting bacteria, Quantitative reverse transcription-PCRAbstract
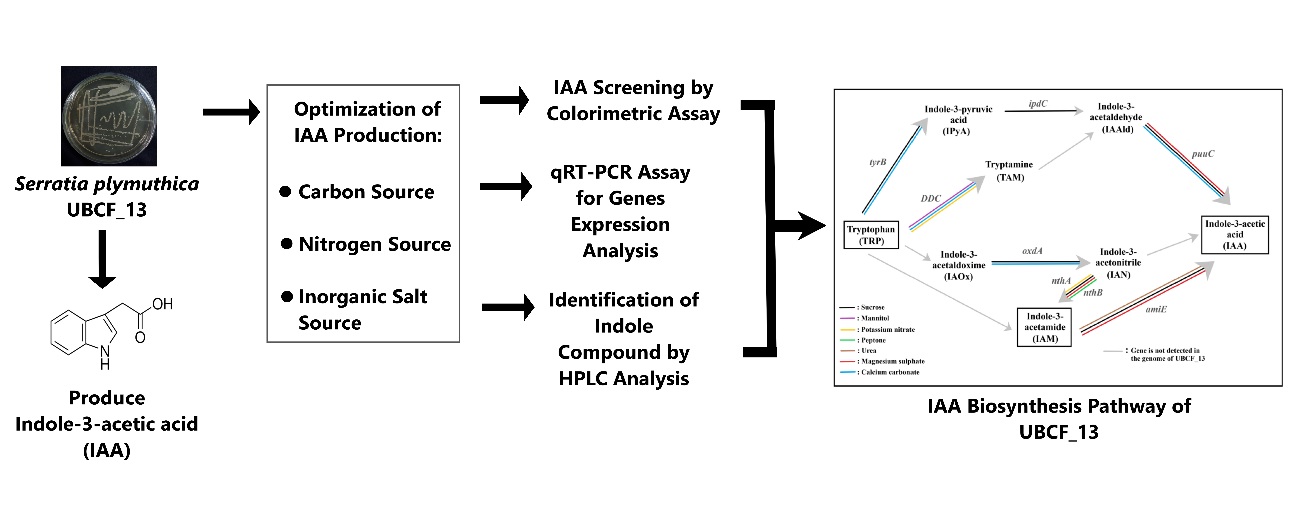
Serratia plymuthica UBCF_13 produces the maximum level of indole-3-acetic acid (IAA) in yeast mannitol medium. However, the impact of medium ingredients on gene expression and metabolites related to IAA synthesis remains unclear. Therefore, Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) was used to assess the effects of culture medium optimization components on the genes involved in IAA production. High-Performance Liquid Chromatography (HPLC) has been employed to investigate the impact of medium constituents on the metabolites associated with the biosynthesis of IAA. The highest IAA production was found in sucrose and yeast extract as the carbon and nitrogen sources (134.6 µg/mL). It was also discovered that calcium carbonate and magnesium sulfate are significant inorganic salts for UBCF_13 in the production of IAA. Most genes showed higher levels of expression when sucrose was used as the carbon source, including ipdC, nthA, puuC, amiE, oxdA, tyrB and nthB. Furthermore, the expression of tyrB, puuC, DDC and oxdA was upregulated in response to calcium carbonate, while puuC, nthA, nthB and amiE expression levels were elevated in the presence of magnesium sulfate. Indole-3-acetamide (IAM) was identified with HPLC as an intermediate product in several optimized culture media, with the highest IAA concentration (429.79 µg/mL) which was observed in yeast sucrose medium. Thus, sucrose played a pivotal role in IAA biosynthesis, and yeast sucrose medium supplemented with complete inorganic salts emerged as the optimal medium for IAA production by UBCF_13.
HIGHLIGHTS
- This is the 1st study to evaluate the effect of each medium culture component on the expression of IAA synthesis genes in bacteria
- The expression of several IAA synthesis genes (amiE, nthA, oxdA, tyrB, nthB, puuC and ipdC) was upregulated by using sucrose as the carbon source
- Magnesium sulphate and calcium carbonate were shown to have a significant influence on IAA production, which also increased the expression of several IAA synthesis genes in UBCF_13
- The increase in the expression of genes linked to the IAM pathway (nthA, nthB and amiE) and the identification of IAM as the only intermediate in the extract of optimum medium culture suggest that UBCF_13 may use the IAM pathway to synthesize IAA
- It was discovered that sucrose and yeast extract with inorganic salts (dipotassium phosphate, magnesium sulphate, sodium chloride and calcium carbonate) were the optimal medium composition to produce IAA by UBCF_13
GRAPHICAL ABSTRACT
Downloads
Metrics
References
K Negacz, P Vellinga, E Barrett-Lennard, R Choukr-Allah and T Elzenga. Future of sustainable agriculture in saline environments. 1st ed. CRC Press, Boca Raton, Florida, 2021.
S Alström and B Gerhardson. Characteristics of Serratia plymuthica isolate from plant rhizospheres. Plant Soil 1987; 103, 185-9.
G Berg. Diversity of antifungal and plant‐associated Serratia plymuthica strains. J. Appl. Microbiol. 2000; 88, 952-60.
DD Vleesschauwer and M Höfte. Using Serratia plymuthica to control fungal pathogens of plants. CABI Reviews 2007. https://doi.org/10.1079/PAVSNNR20072046.
X Liu, Y Wu, Y Chen, F Xu, N Halliday, K Gao, KG Chan and M Cámara. RpoS differentially affects the general stress response and biofilm formation in the endophytic Serratia plymuthica G3. Res. Microbiol. 2016; 167, 168-77.
R Schmidt, VD Jager, D Zühlke, C Wolff, J Bernhardt, K Cankar, J Beekwilder, WV Ijcken, F Sleutels, WD Boer, K Riedel and P Garbeva. Fungal volatile compounds induce production of the secondary metabolite sodorifen in Serratia plymuthica PRI-2C. Sci. Rep. 2017; 7, 862.
MA Matilla, Z Udaondo and GPC Salmond. Genome sequence of the oocydin a-producing rhizobacterium Serratia plymuthica 4Rx5. Microbiol. Resource Announcements 2018; 7, e00997.
S Cleto, GVD Auwera, C Almeida, MJ Vieira, H Vlamakis and R Kolter. Genome sequence of Serratia plymuthica V4. Genome Announcements 2014; 2, e00340-14.
E Adam, H Müller, A Erlacher and G Berg. Complete genome sequences of the Serratia plymuthica strains 3Rp8 and 3Re4-18, two rhizosphere bacteria with antagonistic activity towards fungal phytopathogens and plant growth promoting abilities. Stand. Genomic Sci. 2016; 11, 61.
SR Restrepo, CC Henao, LMA Galvis, JCB Pérez, RAH Sánchez and SDG García. Siderophore containing extract from Serratia plymuthica AED38 as an efficient strategy against avocado root rot caused by Phytophthora cinnamomi. Biocontrol Sci. Tech. 2021; 31, 284-98.
MA Borgi, I Saidi, A Moula, S Rhimi and M Rhimi. The attractive Serratia plymuthica BMA1 strain with high rock phosphate-solubilizing activity and its effect on the growth and phosphorus uptake by Vicia faba L. plants. Geomicrobiol. J. 2020; 37, 437-45.
HT Nguyen, HG Kim, NH Yu, IM Hwang, H Kim, YC Kim and JC Kim. In vitro and in vivo antibacterial activity of serratamid, a novel peptide-polyketide antibiotic isolated from Serratia plymuthica C1, against phytopathogenic bacteria. J. Agr. Food Chem. 2021; 69, 5471-80.
NP Nordstedt and ML Jones. Genomic analysis of Serratia plymuthica MBSA-MJ1: A plant growth promoting rhizobacteria that improves water stress tolerance in greenhouse ornamentals. Front. Microbiol. 2021; 12, 653556.
G Ilangumaran and DL Smith. Plant growth promoting rhizobacteria in amelioration of salinity stress: A systems biology perspective. Front. Plant Sci. 2017; 8, 1768.
SM Kang, R Shahzad, S Bilal, AL Khan, YG Park, KE Lee, S Asaf, MA Khan and IJ Lee. Indole-3-acetic-acid and ACC deaminase producing Leclercia adecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 2019; 19, 80.
A Kumar, S Singh, AK Gaurav, S Srivastava and JP Verma. Plant growth-promoting bacteria: Biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 2020; 11, 1216.
G Mannino, L Nerva, T Gritli, M Novero, V Fiorilli, M Bacem, CM Bertea, E Lumini, W Chitarra and R Balestrini. Effects of different microbial inocula on tomato tolerance to water deficit. Agronomy 2020; 10, 170.
C Petrillo, E Vitale, P Ambrosino, C Arena and R Isticato. Plant growth-promoting bacterial consortia as a strategy to alleviate drought stress in Spinacia oleracea. Microorganisms 2022; 10, 1798.
R Çakmakçı, G Mosber, AH Milton, F Alatürk and B Ali. The effect of auxin and auxin-producing bacteria on the growth, essential oil yield, and composition in medicinal and aromatic plants. Curr. Microbiol. 2020; 77, 564-77.
K Gkarmiri, RD Finlay, S Alström, E Thomas, MA Cubeta and N Högberg. Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica. BMC Genom. 2015; 16, 630.
DN Proença, S Schwab, MS Vidal, JI Baldani, GR Xavier and PV Morais. The nematicide Serratia plymuthica M24T3 colonizes Arabidopsis thaliana, stimulates plant growth, and presents plant beneficial potential. Braz. J. Microbiol. 2019; 50, 777-89.
M Zhang, L Yang, R Hao, X Bai, Y Wang and X Yu. Drought-tolerant plant growth-promoting rhizobacteria isolated from jujube (Ziziphus jujuba) and their potential to enhance drought tolerance. Plant Soil 2020; 452, 423-40.
A Fiodor, N Ajijah, L Dziewit and K Pranaw. Biopriming of seed with plant growth-promoting bacteria for improved germination and seedling growth. Front. Microbiol. 2023; 14, 1142966.
SN Aisyah, J Maldoni, I Sulastri, W Suryati, Y Marlisa, L Herliana, L Syukriani, R Renfiyeni and J Jamsari. Unraveling the optimal culture condition for the antifungal activity and IAA production of phylloplane Serratia plymuthica. Plant Pathol. J. 2019; 18, 31-8.
LA Yusfi, DH Tjong, I Chaniago and J Jamsari. Culture medium optimization for indole-3-acetic acid production by Serratia plymuthica UBCF_13. IOP Conf. Ser. Earth Environ. Sci. 2021; 741, 12059.
LA Yusfi, DH Tjong, I Chaniago, A Salsabilla and J Jamsari. Growth phase influence the gene expression and metabolite production related to indole-3-acetic acid (IAA) biosynthesis by Serratia plymuthica UBCF_13. Pakistan J. Biol. Sci. 2022; 25, 1047-57.
EMT El-Mansi, CFA Bryce, B Dahhou, S Sanchez, AL Demain and AR Allman. Fermentation microbiology and biotechnology. 3rd eds. CRC Press, Boca Raton, 2021.
WH Kampen. Nutritional requirements in fermentation processes. In: Fermentation and biochemical engineering handbook. 3rd eds. William Andrew Publishing, Boston, 2014, p. 37-57.
S Chandra, K Askari and M Kumari. Optimization of indole acetic acid production by isolated bacteria from Stevia rebaudiana rhizosphere and its effects on plant growth. J. Genet. Eng. Biotechnol. 2018; 16, 581-6.
S Ham, H Yoon, JM Park and YG Park. Optimization of fermentation medium for indole acetic acid production by Pseudarthrobacter sp. NIBRBAC000502770. Appl. Biochem. Biotechnol. 2021; 193, 2567-79.
EM Myo, B Ge, J Ma, H Cui, B Liu, L Shi, M Jiang and K Zhang. Indole-3-acetic acid production by Streptomyces fradiae NKZ-259 and its formulation to enhance plant growth. BMC Microbiol. 2019; 19, 155.
S Lebrazi, M Fadil, M Chraibi and K Fikri-Benbrahim. Screening and optimization of indole-3-acetic acid production by Rhizobium sp. strain using response surface methodology. J. Genet. Eng. Biotechnol. 2020; 18, 21.
A Romero-Rodríguez, N Maldonado-Carmona, B Ruiz-Villafán, N Koirala, D Rocha and S Sánchez. Interplay between carbon, nitrogen and phosphate utilization in the control of secondary metabolite production in Streptomyces. Antonie Van Leeuwenhoek 2018; 111, 761-81.
A Smith, A Kaczmar, RA Bamford, C Smith, S Frustaci, A Kovacs-Simon, P O’Neill, K Moore, K Paszkiewicz, RW Titball and S Pagliara. The culture environment influences both gene regulation and phenotypic heterogeneity in Escherichia coli. Front. Microbiol. 2018; 9, 1739.
F Shatila, MM Diallo, U Şahar, G Ozdemir and HT Yalçın. The effect of carbon, nitrogen and iron ions on mono-rhamnolipid production and rhamnolipid synthesis gene expression by Pseudomonas aeruginosa ATCC 15442. Arch. Microbiol. 2020; 202, 1407-17.
EL Imada, AADPRD Santos, ALMD Oliveira, M Hungria and EP Rodrigues. Indole-3-acetic acid production via the indole-3-pyruvate pathway by plant growth promoter Rhizobium tropici CIAT 899 is strongly inhibited by ammonium. Res. Microbiol. 2017; 168, 283-92.
M Li, R Guo, F Yu, X Chen, H Zhao, H Li and J Wu. Indole-3-acetic acid biosynthesis pathways in the plant-beneficial bacterium Arthrobacter pascens ZZ21. Int. J. Mol. Sci. 2018; 19, 443.
BX Zhang, PS Li, YY Wang, JJ Wang, XL Liu, XY Wang and XM Hu. Characterization and synthesis of indole-3-acetic acid in plant growth promoting Enterobacter sp. RSC Adv. 2021; 11, 31601-7.
DR Duca and BR Glick. Indole-3-acetic acid biosynthesis and its regulation in plant-associated bacteria. Appl. Microbiol. Biotechnol. 2020; 104, 8607-19.
SN Aisyah, S Sulastri, R Retmi, RH Yani, E Syafriani, L Syukriani, F Fatchiyah, A Bakhtiar and J Jamsari. Suppression of Colletotrichum gloeosporioides by indigenous phyllobacterium and its compatibility with rhizobacteria. Asian J. Plant Pathol. 2017; 11, 139-47.
SA Gordon and RP Weber. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951; 4, 192-5.
KJ Livak and TD Schmittgen. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods 2001; 25, 402-8.
SL Sun, WL Yang, WW Fang, YX Zhao, L Guo, YJ Dai and I Cann. The plant growth-promoting rhizobacterium Variovorax boronicumulans CGMCC 4969 regulates the level of indole-3-acetic acid synthesized from indole-3-acetonitrile. Appl. Environ. Microbiol. 2018; 84, 298-316.
DW Hutmacher. Polymers from biotechnology. In: Encyclopedia of materials: Science and technology. Elsevier, Oxford, 2001, p. 7680-3.
JN BeMiller. Carbohydrate chemistry for food scientists. 3rd eds. AACC International Press, Minnesota, 2019.
P Nutaratat, W Amsri, N Srisuk, P Arunrattiyakorn and S Limtong. Indole-3-acetic acid production by newly isolated red yeast Rhodosporidium paludigenum. J. Gen. Appl. Microbiol. 2015; 61, 1-9.
S Panigrahi, S Mohanty and CC Rath. Characterization of endophytic bacteria Enterobacter cloacae MG00145 isolated from Ocimum sanctum with Indole Acetic Acid (IAA) production and plant growth promoting capabilities against selected crops. South Afr. J. Bot. 2020; 134, 17-26.
S Rushabh, C Kajal, P Prittesh, N Amaresan and R Krishnamurthy. Isolation, characterization, and optimization of indole acetic acid-producing Providencia species (7MM11) and their effect on tomato (Lycopersicon esculentum) seedlings. Biocatal. Agr. Biotechnol. 2020; 28, 101732.
LT Xa, NK Nghia and HB Tecimen. Environmental factors modulating indole-3-acetic acid biosynthesis by four nitrogen fixing bacteria in a liquid culture medium. Environ. Nat. Resource J. 2022; 20, 279-87.
JM Jeckelmann and B Erni. Transporters of glucose and other carbohydrates in bacteria. Pflugers Arch. 2020; 472, 1129-53.
S Abbasiliasi, JS Tan, TAT Ibrahim, F Bashokouh, NR Ramakrishnan, S Mustafa and AB Ariff. Fermentation factors influencing the production of bacteriocins by lactic acid bacteria: A review. RSC Adv. 2017; 7, 29395-420.
J Barrios-González. Secondary metabolites production: Physiological advantages in solid-state fermentation. In: Current developments in biotechnology and bioengineering. Elsevier, Oxford, 2018, p. 257-83.
P Nutaratat, A Monprasit and N Srisuk. High-yield production of indole-3-acetic acid by Enterobacter sp. DMKU-RP206, a rice phyllosphere bacterium that possesses plant growth-promoting traits. 3 Biotech 2017; 7, 305.
N Bhutani, R Maheshwari, M Negi and P Suneja. Optimization of IAA production by endophytic Bacillus spp. from Vigna radiata for their potential use as plant growth promoters. Isr. J. Plant Sci. 2018; 65, 83-96.
SA Bessai, L Bensidhoum and EH Nabti. Optimization of IAA production by telluric bacteria isolated from northern Algeria. Biocatal. Agr. Biotechnol. 2022; 41, 102319.
T Wang, S Flint and J Palmer. Magnesium and calcium ions: Roles in bacterial cell attachment and biofilm structure maturation. Biofouling 2019; 35, 959-74.
S Harirchi, Z Etemadifar, A Mahboubi, F Yazdian and MJ Taherzadeh. The effect of calcium/magnesium ratio on the biomass production of a novel thermoalkaliphilic Aeribacillus pallidus strain with highly heat-resistant spores. Curr. Microbiol. 2020; 77, 2565-74.
LA Yusfi, DH Tjong, I Chaniago and J Jamsari. Screening the effect of YM media component and tryptophan levels on IAA production of Serratia plymuthica UBCF_13. AIP Conf. Proc. 2023; 2730, 80001.
G Joshi, V Kumar and SK Brahmachari. Screening and identification of novel halotolerant bacterial strains and assessment for insoluble phosphate solubilization and IAA production. Bull. Natl. Res. Cent. 2021; 45, 83.
LG Sarmiento-López, M López-Meyer, IE Maldonado-Mendoza, FR Quiroz-Figueroa, GG Sepúlveda-Jiménez and M Rodríguez-Monroy. Production of indole-3-acetic acid by Bacillus circulans E9 in a low-cost medium in a bioreactor. J. Biosci. Bioeng. 2022; 134, 21-8.
K Allali, M Zamoum, A Benadjila, A Zitouni and Y Goudjal. Valorization of tomato plant wastes and optimization of growth conditions for indole-3-acetic acid production by Streptomyces plicatus strain PT2: Actinobacteria and phytohormones. J. Microbiol. Biotechnol. Food Sci. 2023; 13, e9580.
MMA Elsoud, SF Hasan and MM Elhateir. Optimization of Indole-3-acetic acid production by Bacillus velezensis isolated from Pyrus rhizosphere and its effect on plant growth. Biocatal. Agr. Biotechnol. 2023; 50, 102714.
HJ Shin, S Woo, GY Jung and JM Park. Indole-3-acetic acid production from alginate by Vibrio sp. dhg: Physiology and characteristics. Biotechnol. Bioprocess Eng. 2023; 28, 695-703.
Q Wang and CT Nomura. Monitoring differences in gene expression levels and polyhydroxyalkanoate (PHA) production in Pseudomonas putida KT2440 grown on different carbon sources. J. Biosci. Bioeng. 2010; 110, 653-59.
L Chen, Y Luo, H Wang, S Liu, Y Shen and M Wang. Effects of Glucose and starch on lactate production by newly isolated Streptococcus bovis S1 from Saanen goats. Appl. Environ. Microbiol. 2016; 82, 5982-9.
C Hamilton, M Calusinska, S Baptiste, J Masset, L Beckers, P Thonart and S Hiligsmann. Effect of the nitrogen source on the hydrogen production metabolism and hydrogenases of Clostridium butyricum CWBI1009. Int. J. Hydrogen Energ. 2018; 43, 5451-62.
MJD Valle, JE Laiño, GSD Giori and JG LeBlanc. Factors stimulating riboflavin produced by Lactobacillus plantarum CRL 725 grown in a semi-defined medium. J. Basic Microbiol. 2017; 57, 245-52.
J Liu, H Li, X Zhang, L Yue, W Lu, S Ma, Z Zhu, D Wang, H Zhu and J Wang. Cost-efficient production of the sphingan WL gum by Sphingomonas sp. WG using molasses and sucrose as the carbon sources. Mar. Biotechnol. 2023; 25, 192-203.
M Szkop and W Bielawski. TyrB-2 and phhC genes of Pseudomonas putida encode aromatic amino acid aminotransferase isozymes: Evidence at the protein level. Amino Acids 2013; 45, 351-8.
H Kawaguchi, H Miyagawa, S Nakamura-Tsuruta, N Takaya, C Ogino and A Kondo. Enhanced phenyllactic acid production in Escherichia coli via oxygen limitation and shikimate pathway gene expression. Biotechnol. J. 2019; 14, e1800478.
JA Barbosa-Nuñez, OA Palacios, LE De-Bashan, R Snell-Castro, RI Corona-González and FJ Choix. Active indole‐3‐acetic acid biosynthesis by the bacterium Azospirillum brasilense cultured under a biogas atmosphere enables its beneficial association with microalgae. J. Appl. Microbiol. 2022; 132, 3650-63.
M Li, T Li, M Zhou, M Li, Y Zhao, J Xu, F Hu and H Li. Caenorhabditis elegans extracts stimulate IAA biosynthesis in Arthrobacter pascens ZZ21 via the indole-3-pyruvic acid pathway. Microorganisms 2021; 9, 970.
H Wu, J Yang, P Shen, Q Li, W Wu, X Jiang, L Qin, J Huang, X Cao and F Qi. High-level production of indole-3-acetic acid in the metabolically engineered Escherichia coli. J. Agr. Food Chem. 2021; 69, 1916-24.
LD Tullio, AS Nakatani, DF Gomes, FJ Ollero, M Megías and M Hungria. Revealing the roles of y4wF and tidC genes in Rhizobium tropici CIAT 899: Biosynthesis of indolic compounds and impact on symbiotic properties. Arch. Microbiol. 2019; 201, 171-83.
J Buezo, R Esteban, A Cornejo, P López-Gómez, D Marino, A Chamizo-Ampudia, MJ Gil, V Martínez-Merino and JF Moran. IAOx induces the SUR phenotype and differential signalling from IAA under different types of nitrogen nutrition in Medicago truncatula roots. Plant Sci. 2019; 287, 110176.
CL Patten, AJC Blakney and TJD Coulson. Activity, distribution and function of indole-3-acetic acid biosynthetic pathways in bacteria. Crit. Rev. Microbiol. 2013; 39, 395-415.
Y Liu, X Jiang, D Guan, W Zhou, M Ma, B Zhao, F Cao, L Li and J Li. Transcriptional analysis of genes involved in competitive nodulation in Bradyrhizobium diazoefficiens at the presence of soybean root exudates. Sci. Rep. 2017; 7, 10946.
D Duca, DR Rose and BR Glick. Characterization of a nitrilase and a nitrile hydratase from Pseudomonas sp. strain UW4 that converts indole-3-acetonitrile to indole-3-acetic acid. Appl. Environ. Microbiol. 2014; 80, 4640-9.
CEM Grossi, E Fantino, F Serral, MS Zawoznik, DAFD Porto and RM Ulloa. Methylobacterium sp. 2A is a plant growth-promoting rhizobacteria that has the potential to improve potato crop yield under adverse conditions. Front. Plant Sci. 2020; 11, 71.
FP Matteoli, H Passarelli-Araujo, RJA Reis, LOD Rocha, EMD Souza, L Aravind, FL Olivares and TM Venancio. Genome sequencing and assessment of plant growth-promoting properties of a Serratia marcescens strain isolated from vermicompost. BMC Genom. 2018; 19, 750.
L Ouyang, H Pei and Z Xu. Low nitrogen stress stimulating the indole-3-acetic acid biosynthesis of Serratia sp. ZM is vital for the survival of the bacterium and its plant growth-promoting characteristic. Arch. Microbiol. 2017; 199, 425-32.
R Kotoky and P Pandey. Rhizosphere assisted biodegradation of benzo(a)pyrene by cadmium resistant plant-probiotic Serratia marcescens S2I7, and its genomic traits. Sci. Rep. 2020; 10, 5279.
BK Jung, AR Khan, SJ Hong, GS Park, YJ Park, CE Park, HJ Jeon, SE Lee and JH Shin. Genomic and phenotypic analyses of Serratia fonticola strain GS2: A rhizobacterium isolated from sesame rhizosphere that promotes plant growth and produces N-acyl homoserine lactone. J. Biotechnol. 2017; 241, 158-62.
AR Khan, GS Park, S Asaf, SJ Hong, BK Jung and JH Shin. Complete genome analysis of Serratia marcescens RSC-14: A plant growth-promoting bacterium that alleviates cadmium stress in host plants. PLoS One 2017; 12, e0171534.
MA Hassan, K Al-Sakkaf, MRS Mohammed, A Dallol, J Al-Maghrabi, A Aldahlawi, S Ashoor, M Maamra, J Ragoussis, W Wu, MI Khan, AL Al-Malki and H Choudhry. Integration of transcriptome and metabolome provides unique insights to pathways associated with obese breast cancer patients. Front. Oncol. 2020; 10, 804.
Q Nong, MK Malviya, MK Solanki, L Lin, J Xie, Z Mo, Z Wang, X Song, X Huang, C Li and Y Li. Integrated metabolomic and transcriptomic study unveils the gene regulatory mechanisms of sugarcane growth promotion during interaction with an endophytic nitrogen-fixing bacteria. BMC Plant Biol. 2023; 23, 54.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.