Isolation of Genes Encoding for Human Epidermal Growth Factor (hEGF) from Human Blood: In Vitro and In Silico Study
DOI:
https://doi.org/10.48048/tis.2023.6563Keywords:
Docking, DNA isolation, hEGF, Phylogenetics, Protein modelingAbstract
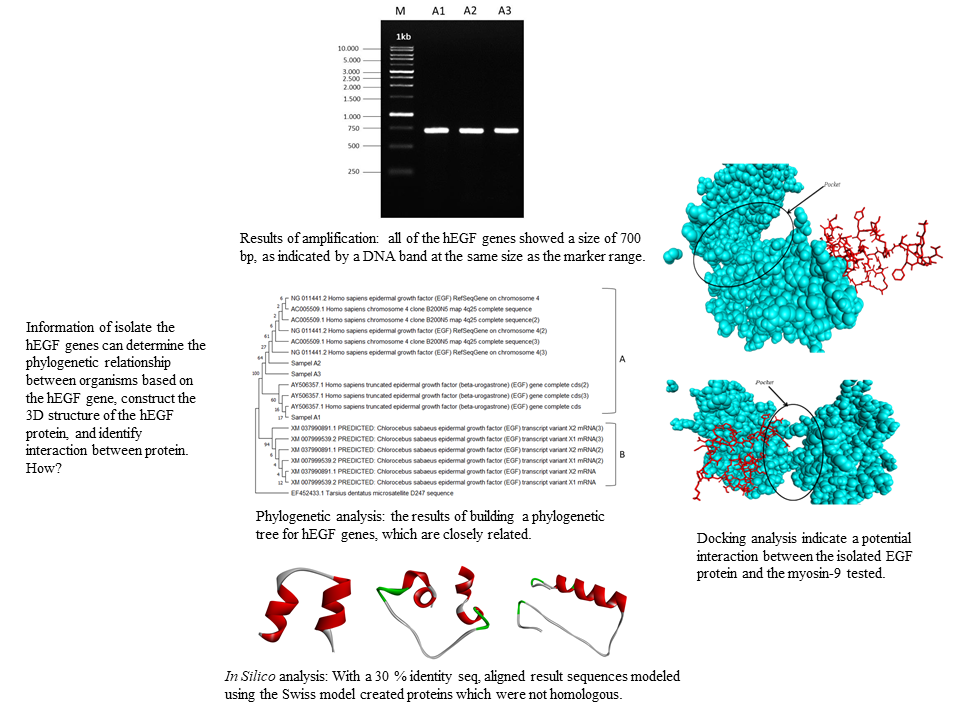
The 6.12 kDa human epidermal growth factor (hEGF) protein is expressed in the body and plays a role in cell proliferation and differentiation. This research aims to isolate the hEGF genes, determine the phylogenetic relationship between organisms based on the hEGF gene, construct the 3D structure of the hEGF protein, and identify the precise binding position between the hEGF protein and the tested Myosin-9. DNA was isolated from blood of healthy individuals followed by amplification using specified primers. DNA sequencing was utilized to identify the amplification results before generating the phylogenetic trees. The retrieved sequences were then modeled using the Swiss model and docking proteins in silico. In three samples, DNA was successfully amplified to 700 bp and the phylogenetic analysis revealed that the three hEGF gene samples belonged to the same clades. With a 30 % identity seq, aligned result sequences modeled using the Swiss model created proteins which were not homologous. The outcomes of this modeling were compared to the hEGF protein from the database, which had a sequence identity of greater than 90 %. The i-RMSD value of the target protein (5.4 +/- 0.4), the Van der Waals energy (-29.3 +/- 1.7), and the Z-score were determined by simulation (-1.7). These results indicate a potential interaction between the isolated EGF protein and the myosin-9 tested.
HIGHLIGHTS
- Human Epidermal Growth Factor (hEGF) protein is a potential protein expressed in the body and plays a role in cell proliferation and differentiation
- Isolation of the EGF gene through human blood can be carried out to get data on protein size, phylogenetic analysis, and protein structure
- The results of in vitro study were modeled in silico to obtain the three-dimensional structure as well as intermolecular interactions of the protein
- Information beginning this study is used in follow-up research to predict EGF communications with other molecules
GRAPHICAL ABSTRACT
Downloads
Metrics
References
K Vinay, A K Abbas, N Fausto and JC Aster. Robbins and Cotran pathologic basis of disease, professional edition e-book. Elsevier health sciences, Canada, 2014, p. 338-341.
Epidermal Growth Factor, available at: https://www.ncbi.nlm.nih.gov, accessed December 2022.
Y Kang, W He, C Ren, J Qiao, Q Guo, J Hu and L Wang. Advances in targeted therapy mainly based on signal pathways for nasopharyngeal carcinoma. Signal Transduct. Targeted Ther. 2020; 5, 1-20.
J Zhu, G Jiang, W Hong, Y Zhang, B Xu, G Song and L Ruan. Rapid gelation of oxidized hyaluronic acid and succinyl chitosan for integration with insulin-loaded micelles and epidermal growth factor on diabetic wound healing. Mater. Sci. Eng. 2020; 117, 1-12.
FD Crendhuty and S Megantara. Sediaan Hydrogel mengandung Epidermal Growth Factor dalam Penyembuhan Luka. Farmaka 2019; 17, 410-6.
S Pouranvari, F Ebrahimi, G Javadi and B Maddah. Cloning, expression, and cost effective purification of authentic human epidermal growth factor with high activity. Iranian Red Crescent Med. J. 2016; 18, 1-8.
X Zheng, X Wu, X Fu, D Dai and F Wang. Expression and purification of human Epidermal Growth Factor (hEGF) fused with GB1. Biotechnol. Biotechnol. Equip. 2016; 30, 813-8.
GA Sandag and SW Taju. Bioinformatics tools for data processing and prediction of protein function. CogITo Smart J. 2018; 4, 305-15.
R Solfaine, L Muniroh and WW Mubarokah. Study of doking moleculer flavonoid Coleus amboinicus in TGF-1b receptor and lowering MDA concentration on Cisplatin-induce Wistar Rats. Jurnal Sains Veteriner 2020; 38, 151-8.
R Prasetiawati, M Suherman, B Permana and R Rahmawati. Molecular docking study of anthocyanidin compounds against Epidermal Growth Factor Receptor (EGFR) as anti-lung cancer. Indonesian J. Pharmaceut. Sci. Tech. 2021; 8, 8-20.
S Suherlan, R Rohayah and TM Fakih. Uji aktivias antikanker payudara senyawa andrografolida dari tumbuhan sambiloto (Andrographis paniculata (Burm F) Ness.) terhadap human epidermal growth factor receptor 2 (HER-2) secara in silico. Jurnal Ilmiah Farmasi Farmasyifa 2021, 4, 39-50.
M Malek, F Mashayekhi and Z Salehi. Epidermal growth factor +61A/G (rs4444903) promoter polymorphism and serum levels are linked to idiopathic male infertility. British J. Biomed. Sci. 2020; 78, 92-4.
S Siswandono. Pengembangan Obat Baru, Cetakan I. Airlangga University Press, Surabaya, 2014, p. 59-60.
T Pervaiz, X Sun, Y Zhang, R Tao, J Zhang and J Fang. Association between chloroplast and mitochondrial DNA sequences in Chinese Prunus genotypes (Prunus persica, Prunus domestica, and Prunus avium). BMC Plant Biol. 2015; 15, 1-10.
G Koetsir and C Eric. A practical guide to analyzing nucleic acid concentration and purify with microvolume spectrophotometers. New England BioLabs 2019; 7, 1-9.
M Ghaheri, D Kahrizi, K Yari, A Babaie, RS Suthar and E Kazemi. A comparative evaluation of four DNA extraction protocols from whole blood sample. Cellular Mol. Biol. 2016; 62, 120-4.
M Mitra. DNA sequencing basics and its applications. SCIOL Genet Sci. 2018; 1, 80-4.
DW Ardiana. Teknik isolasi DNA genom tanaman pepaya dan jeruk dengan menggunakan modifikasi bufer CTAB. Buletin Teknik Pertanian 2009; 14, 12-6.
Customized Products. Available at: https://base-asia.com/wp-content/uploads/2021, accessed November 2022.
W Windasari, MF Islam and A Taher. Analisis Keragaman Genetik Daerah 3’utr Gen Reseptor Lipoprotein Densitas Rendah (LDLR) Dari Individu Asal Papua. In: Prosiding Seminar Nasional MIPA UNIPA, Universitas Papua, Manokwari. 2022, p. 157-63.
JG Rohwer. Toward a phylogenetic classification of the Lauraceae: Evidence from matK sequences. Syst. Bot. 2000; 25, 60-71.
R Widayanti and S Trini. Studi keragaman genetik Tarsius sp. asal kalimantan, sumatera, dan sulawesi berdasarkan sekuen gen NADH dehidrogenase sub-unit 4L (ND4L). Jurnal Kedokteran Hewan. 2012; 6, 105-11.
R Widayanti, S Trini and TA Wayan. Keragaman genetik gen NADH dehydrogenase subunit 6 pada monyet hantu (Tarsius Sp.). Jurnal Veteriner. 2013; 14, 239-49.
AS Darupamenang, JK Beivy, SA Nio and ET Trina. Analisis Filigenetik Genus Alocasia. Jurnal Bios Logos 2022; 12, 157-63.
AR Simbolon, M Ompi, E Widyastuti and DA Wulandari. Penggunaan DNA barcoding dalam mengidentifikasi larva gastropoda (family cymatiidae) di perairan kepulauan sangihe-talaud, sulawesi utara. Bawal Widya Riset Perikanan Tangkap. 2021; 13, 145-55.
N Komari, S Hadi and E Suhartono. Pemodelan Protein dengan Homology Modeling menggunakan SWISS-MODEL: Protein Modeling with Homology Modeling using SWISS-MODEL. Jurnal Jejaring Matematika dan Sains 2020; 2, 65-7.
F Kiefer, K Arnold, M Künzli, L Bordoli and T Schwede. The swiss-model repository and associated resources. Nucleic Acids Res. 2009; 37, 387-92.
FZ Saudale. Pemodelan Homologi komparatif struktur 3d protein dalam desain dan pengembangan obat. Al-Kimia 2020; 8, 1-11.
S Suprianto and IM Budiarsa. Analisis in silico protein clock (circadian locomotor output cycles kaput) pada Patagioenas fasciata monilis. Jurnal Sains dan Teknologi 2021; 10, 9-15.
Y Yeni and DH Tjahjono. Homology modeling epitop isocitrate dehydrogenase tipe 1 (r132h) menggunakan modeller, i-tasser dan (Ps) 2 untuk vaksin glioma. Farmasains: Jurnal Ilmiah Ilmu Kefarmasian 2017; 4, 21-32.
R Chen. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol. Adv. 2012; 30. 1102.
V Charitou, SCV Keulen and AM Bonvin. Cyclization and docking protocol for cyclic peptide-protein modeling using HADDOCK2.4. J. Chem. Theor. Comput. 2022; 18, 4027-40.
A Duelli, B Kiss, I Lundholm, A Bodor, MV Petoukhov, DI Svergun, N Latzio and G Katona. The C-terminal random coil region tunes the Ca2+-binding affinity of S100A4 through conformational activation. PloS One 2014; 9, 97654.
FZ Muttaqin, H Ismail and Muhammad. Studi molecular docking, molecular dynamic, dan prediksi toksisitas senyawa turunan alkaloid naftiridin sebagai inhibitor protein kasein kinase 2-Α pada kanker leukemia. J. Pharmacoscript 2019; 2. 49-64.
MF Lensink, N Nadzirin, S Velankar and SJ Wodak. Modeling protein‐protein, protein‐peptide, and protein‐oligosaccharide complexes: CAPRI 7th edition. Proteins Struct. Funct. Bioinform. 2020; 88, 916-38.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






