Bioprospecting of Rhizobia as Plant Growth Promoting Rhizobacteria Potential from Root Nodules of Groundnut (Arachis hypogaea L.)
DOI:
https://doi.org/10.48048/tis.2024.7651Keywords:
Arachis hypogaea, IAA synthesize, Nitrogen fixation, Proteases, Phosphate solubilization, Rhizobium sp.Abstract
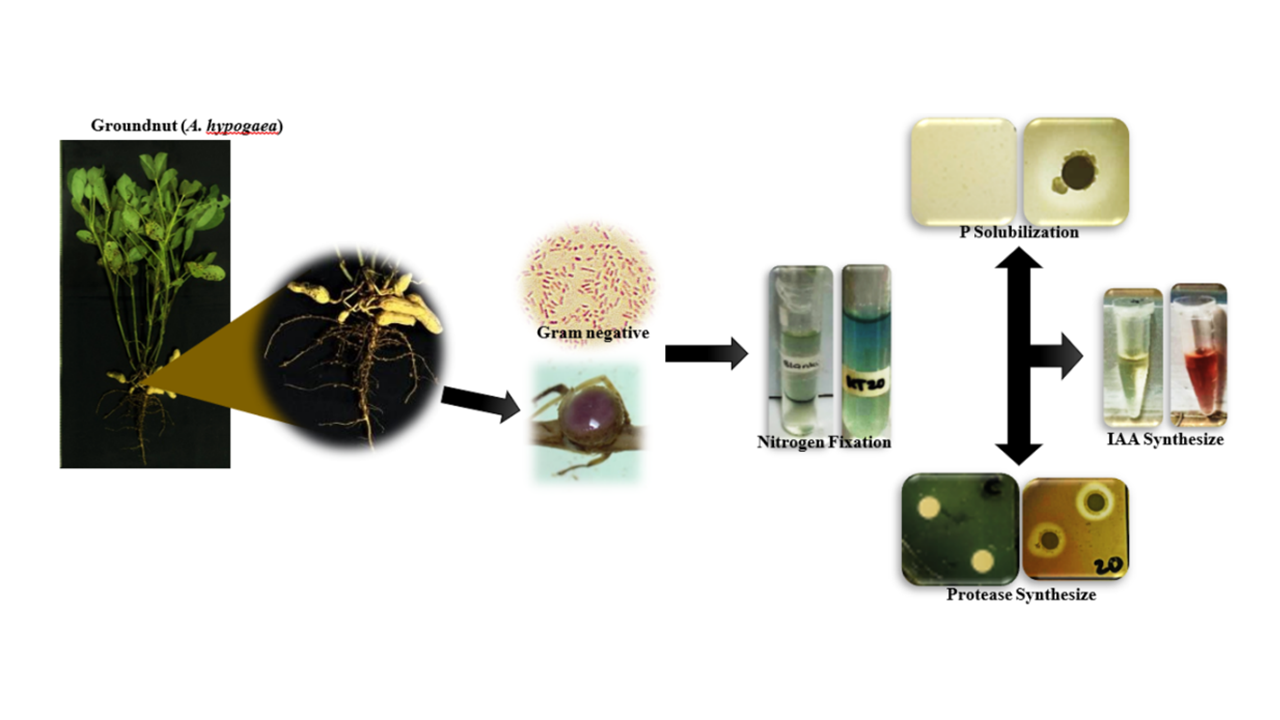
Rhizobia are bacteria that symbiosis with host plant, as shown in the root nodules formation, and provide nitrogen that can be absorbed by plants in greater quantities than rhizobacteria. Available Nitrogen, which absorbed by plants, is the essential requirement for plant growth because its role in increasing yield and quality, hence it is needed in greater quantities than other nutrients. The study aimed to determine the macroscopic and microscopic diversity of rhizobia isolates from the groundnut nodules and their potential as PGPRs, and to identify 16S rRNA isolates with the best potential as PGPRs molecularly. The methods used were isolation from root nodules, screening of PGPR potential, molecular identification based on the 16S rRNA gene, and phylogenetic analysis to determine their kinship. Based on the isolation results, 17 Gram-negative isolates were obtained white to pink or orange color on AG media with various colony characteristics in terms of shape, margin, elevation, and texture. KT 20, which was selected as rhizobia isolate with the best potential as PGPR, has ammonium concentration of 23.12 ppm, synthesizes IAA with a concentration of 10.36 ppm, and phosphates solubilization activity, although its ability to synthesize proteases is low. The results of molecular identification of 16S rRNA showed that KT 20 belongs to the Rhizobium genus with a similarity of 99.48 % and bootstrap value of 96 %.
HIGHLIGHTS
- Isolate KT 20 (identified rhizobia) has excellent ability in fixing nitrogen, which plants needs the most and legumes requires in large number
- Beside the ability of Isolate KT 20 to fix nitrogen, it also able to synthesize IAA, solubilize phosphate, and synthesize protease
- The ability of isolate KT 20 as PGPR was equal or better than other rhizobacteria that has been analyzed in other in vitro studies
GRAPHICAL ABSTRACT

Downloads
References
S Batool and A Iqbal. Phosphate solubilizing rhizobacteria as alternative of chemical fertilizer for growth and yield of Triticum aestivum (Var. Galaxy 2013). Saudi J. Biol. Sci. 2019; 26, 1400-10.
W Elhaissoufi, S Khourchi, A Ibnyasser, C Ghoulam, Z Rchiad, Y Zeroual, K Lyamlouli and A Bargaz. Phosphate solubilizing rhizobacteria could have a stronger influence on wheat root traits and aboveground physiology than rhizosphere P solubilization. Front. Plant Sci. 2020; 11, 979.
S Ali, S Hameed, M Shahid, M Iqbal, G Lazarovits and A Imran. Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol. Res. 2020; 232, 126389.
PMD Lajudie, M Andrews, J Ardley, B Eardly, E Jumas-Bilak, N Kuzmanović, F Lassalle, K Lindström, R Mhamdi, E Martínez-Romero, L Moulin, SA Mousavi, X Nesme, A Peix, J Puławska, E Steenkamp, T Stępkowski, CF Tian, P Vinuesa, G Wei1, ..., P Young. Minimal standards for the description of new genera and species of rhizobia and agrobacteria. Int. J. Syst. Evol. Microbiol. 2019; 69, 1852-63.
Y Yanti, FF Astuti, T Habazar and CR Nasution. Screening of rhizobacteria from rhizosphere of healthy chili to control bacterial wilt disease and to promote growth and yield of chili. Biodiversitas J. Biol. Divers. 2016; 18, 1-9.
Q Wang, J Liu and H Zhu. Genetic and molecular mechanisms underlying symbiotic specificity in legume-rhizobium interactions. Front. Plant Sci. 2018; 9, 313.
MK Meghvansi, K Prasad and SK Mahna. Symbiotic potential, competitiveness and compatibility of indigenous Bradyrhizobium japonicum isolates to three soybean genotypes of two distinct agro-climatic regions of Rajasthan, India. Saudi J. Biol. Sci. 2010; 17, 303-10.
B Sinare, A Miningou, B Nebié, J Eleblu, O Kwadwo, A Traoré, B Zagre and H Desmae. Participatory analysis of groundnut (Arachis hypogaea L.) cropping system and production constraints in Burkina Faso. J. Ethnobiology Ethnomedicine 2021; 17, 2.
S Wanget, M Morales-Corts, R Pérez-Sánchez, N Rostini, M Gómez-Sánchez and A Karuniawan. Agro-morphological and chemical characterization of traditional Indonesian peanut (Arachis hypogaea L.) cultivars. Genetika 2019; 51, 179-98.
T Okubo, S Fukushima and K Minamisawa. Evolution of bradyrhizobium-aeschynomene mutualism: Living testimony of the ancient world or highly evolved state? Plant Cell Phys. 2012; 53, 2000-7.
EC Cocking, SL Kothari, CA Batchelor, S Jain, G Webster, J Jones, J Jotham and MR Davey. Interaction of rhizobia with non-legume crops for symbiotic nitrogen fixation nodulation. Azospirillum VI and Related Microorganisms. NATO ASI Series, Marktoberdorf, Germany, 1995, p. 197-205.
S Goswami, K Raval, Anjana and P Bhat. Bioprospecting of microorganism‐based industrial molecules. In: SP Singh and SK Upadhyay (Eds.). Wiley, New Jersey, 2020.
N Triwahyuningsih and TB Kusmiyarti. Isolation, characterization, and inoculum formulation of nodule forming bacteria of kudzu (Pueraria phaseoloides (Roxb.) Benth.). Int. J. Biosci. Biotechnol. 2018; 6, 10-24.
J Pang, M Palmer, HJ Sun, CO Seymour, L Zhang, BP Hedlund and F Zeng. Diversity of root nodule-associated bacteria of diverse legumes along an elevation gradient in the Kunlun Mountains, China. Front. Microbiol. 2021; 12, 633141.
D Parks, C Peterson and WS Chang. Identification and validation of reference genes for expression analysis in nitrogen-fixing bacteria under environmental stress. Life 2022; 12, 1379.
R Soares, J Trejo, MJ Lorite, E Figueira, J Sanjuán and IVE Castro. Diversity, phylogeny and plant growth promotion traits of nodule associated bacteria isolated from Lotus parviflorus. Microorganisms 2020; 8, 499.
DK Ondieki, EN Nyaboga, JM Wagacha and FB Mwaura. Morphological and genetic diversity of rhizobia nodulating cowpea (Vigna unguiculata L.) from agricultural soils of lower eastern kenya. Int. J. Microbiol. 2017; 2017, 8684921.
Y Thairu, Y Usman and I Nasir. Laboratory perspective of gram staining and its significance in investigations of infectious diseases. Sub Saharan Afr. J. Med. 2014; 1, 168.
JM Day and J Döbereiner. Physiological aspects of N2-fixation by a Spirillum from Digitaria roots. Soil Biol. Biochem. 1976; 8, 45-50.
A Cordova-Rodriguez, ME Rentería-Martínez, CA López-Miranda, JM Guzmán-Ortíz and SF Moreno-Salazar. Simple and sensitive spectrophotometric method for estimating the nitrogen-fixing capacity of bacterial cultures. MethodsX 2022; 9, 101917.
R Pikovskaya. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 1948; 17, 362-70.
S Dhull, R Gera, HS Sheoran and R Kakar. Phosphate solubilization activity of rhizobial strains isolated from root nodule of cluster bean plant native to indian soils. Int. J. Curr. Microbiol. Appl. Sci. 2018; 7, 255-66
H Halimursyadah, Syamsuddin, Nurhayati and D Rizva. Exploration, isolation and characterization of indigenous rhizobacteria from patchouli rhizosphere as PGPR candidates in producing IAA and solubilizing phosphate. IOP Conf. Ser. Earth Environ. Sci. 2022l; 951, 012055.
M Sarwar and RJ Kremer. Determination of bacterially derived auxins using a microplate method. Lett. Appl. Microbiol. 1995; 20, 282-5.
G Pant and PK Agrawal. Isolation and characterization of indole acetic acid producing plant growth promoting rhizobacteria from rhizospheric soil of withania somnifera. J. Biol. Sci. Opin. 2014; 2, 377-83.
VJ Mehta, JT Thumarand SP Singh. Production of alkaline protease from an alkaliphilic actinomycete. Bioresource Tech. 2006; 97, 1650-4
V Kumar, N Sharma and MA Kuchay. Diversity and characterization of plant growth promoting rhizobacteria associated with wheat (Triticum aestivum) rhizosphere in north-western himalayan state of himachal pradesh in India. Int. J. Curr. Microbiol. Appl. Sci. 2020; 9, 747-64.
G Lim, TK Tan and NA Rahim. Variations in amylase and protease activities amongRhizopus isolates. MIRCEN J. Appl. Mmicrobiol. Biotechnol. 1987; 3, 319-22.
I Dewiyanti, D Darmawi, ZA Muchlisin, TZ Helmi, II Arisa, R Rahmiati and E Destri. Cellulase enzyme activity of the bacteria isolated from mangrove ecosystem in Aceh Besar and Banda Aceh. IOP Conf. Ser. Earth Environ. Sci. 2022; 951, 012113.
GNK Raissa, K Ibrahim, T Kaoutar, B Issouf, BGA Maxwell, AD Sélastique and FM Abdelkarim. Molecular and symbiotic efficiency characterization of rhizobia nodulating bambara groundnut (Vigna subterranea L.) from agricultural soils of daloa localities in Côte D’ivoire. Int. J. Curr. Microbiol. Appl. Sci. 2020; 9, 507-19.
GP Telles, GS Araújo, MEMT Walter, MM Brigido and NF Almeida. Live neighbor-joining. BMC Bioinformatics 2018; 19, 172.
FA Azizan, N Roslan, R Ruslan, AA Aznan and AZM Yusoff. Soil NPK variability mapping for harumanis mango grown in greenhouse at Perlis, Malaysia. Int. J. Adv. Sci. Eng. Inform. Tech. 2019; 9, 495-501.
CA Friel. and ML Friesen. Legumes modulate allocation to rhizobial nitrogen fixation in response to factorial light and nitrogen manipulation. Fron. Plant Sci. 2019; 10, 1316.
M. H Lin, PM Gresshoff and BJ Ferguson. Systemic regulation of soybean nodulation by acidic growth conditions. Plant Phys. 2012; 160, 2028-39.
R Kapembwa, AM Mweetwa, M Ngulube and J Yengw. Morphological and biochemical characterization of soybean nodulating rhizobia indigenous to Zambia. Sustain. Agr. Res. 2016; 5, 84.
A Shankar and V Prasad. Potential of desiccation-tolerant plant growth-promoting rhizobacteria in growth augmentation of wheat (Triticum aestivum L.) under drought stress. Front. Microbiol. 2023; 14, 1017167.
C Odori, J Ngaira, J Kinyua and EN Nyaboga. Morphological, genetic diversity and symbiotic functioning of rhizobia isolates nodulating cowpea (Vigna unguiculata L. Walp) in soils of Western Kenya and their tolerance to abiotic stress. Cogent Food Agr. 2020; 6, 1853009.
M Janczarek and A Skorupska. The Rhizobium leguminosarum bv. trifolii RosR: Transcriptional regulator involved in exopolysaccharide production. Mol. Plan Microbe Interac. 2007; 20, 867-81.
H Etesami. Root nodules of legumes: A suitable ecological niche for isolating non-rhizobial bacteria with biotechnological potential in agriculture. Curr. Res. Biotechnol. 2022; 4, 78-86.
P Piromyou, B Buranabanyat, P Tantasawat, P Tittabutr, N Boonkerd and N Teaumroong. Effect of plant growth promoting rhizobacteria (PGPR) inoculation on microbial community structure in rhizosphere of forage corn cultivated in Thailand. Eur. J. Soil Biol. 2011; 47, 44-54.
X Han, K Wu, X Fu and Q Liu. Improving coordination of plant growth and nitrogen metabolism for sustainable agriculture. aBIOTECH 2020; 1, 255-75.
T Shimada and T Hasegawa. Determination of equilibrium structures of bromothymol blue revealed by using quantum chemistry with an aid of multivariate analysis of electronic absorption spectra. Spectrochim. Acta Mol. Biomol. Spectros. 2017; 185, 104-10.
MM Joe, S Deivaraj, A Benson, AJ Henry and G Narendrakumar. Soil extract calcium phosphate media for screening of phosphate-solubilizing bacteria. Agr. Nat. Res. 2018; 52, 305-8.
A Kumar, A Kumar and H Patel. Role of microbes in phosphorus availability and acquisition by plants. Int. J. Curr. Microbiol. Appl. Sci. 2018; 7, 1344-7.
S Gang, S Sharma, M Saraf, M Buck and J Schumacher. Analysis of indole-3-acetic acid (IAA) production in Klebsiella by LC-MS/MS and the Salkowski method. Bio Protocol 2019; 9, e3230.
S Chandra, K Askari and M Kumari. Optimization of indole acetic acid production by isolated bacteria from Stevia rebaudiana rhizosphere and its effects on plant growth. J. Genet. Eng. Biotechnol. 2018; 16, 581-6.
AR Ramadhan, Z Bachruddin, Widodo, Y Erwanto and C Hanim. Isolation and selection of proteolytic lactic acid bacteria from colostrum of dairy cattle. IOP Conf. Ser. Earth Environ. Sci. 2021; 788, 012077.
MA Bhat, V Kumar, MA Bhat, IA Wani, FL Dar, I Farooq, F Bhatti, R Koser, S Rahman and AT Jan. Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (pgpr) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 2020; 11, 1952.
AA Shoukry, HH El-Sebaay and AE El-Ghomary. Assessment of indole acetic acid production from Rhizobium leguminosarum strains. Curr. Sci. Int. 2018; 7, 60-9.
Z Simon, K Mtei, A Gessesse and PA Ndakidemi. Isolation and characterization of nitrogen fixing rhizobia from cultivated and uncultivated soils of Northern Tanzania. Am. J. Plant Sci. 2014; 5, 4050-67.
RI Kabir, S Yeasmin, AKMM Islam and MAR Sarkar. Effect of phosphorus, calcium and boron on the growth and yield of groundnut (Arachis hypogaea L.). Int. J. Bio Sci. Bio Tech. 2013; 5, 51-60.
H Xu, M Detto, S Fang, RL Chazdon, Y Li, BCH Hau, GA Fischer, GD Weiblen, JA Hogan, JK Zimmerman, M Uriarte, J Thompson, J Lian, K Cao, D Kenfack, A Alonso, P Bissiengou, HR Memiaghe, R Valencia, SL Yap, ..., TL Yao. Soil nitrogen concentration mediates the relationship between leguminous trees and neighbor diversity in tropical forests. Comm. Biol. 2020; 3, 317.
G Lucena-Aguilar, AM Sánchez-López, C Barberán-Aceituno, JA Carrillo-Ávila, JA López-Guerrero, and R Aguilar-Quesada. DNA source selection for downstream applications based on dna quality indicators analysis. Biopreservation Biobanking 2016; 14, 264-70.
F Multinu, SC Harrington, J Chen, PR Jeraldo, S Johnson, N Chia and MR Walther-Antonio. Systematic bias introduced by genomic DNA template dilution in 16S rRNA gene-targeted microbiota profiling in human stool homogenates. mSphere 2018; 3, e00560-17.
I Abellan-Schneyder, MS Matchado, S Reitmeier, A Sommer, Z Sewald, J Baumbach, M List and K Neuhaus. Primer, pipelines, parameters: Issues in 16S rRNA gene sequencing. mSphere 2021; 6, e01202-20.
GC Baker, JJ Smith and DA Cowan. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Meth. 2003; 55, 541-55.
KC Samal, JP Sahoo, L Behera and T Dash. Understanding the BLAST (Basic Local Alignment Search Tool) program and step-by-step guide for its use in life science research. Bhartiya Krishi Anusandhan Patrika 2021; 36, 55-61.
C Karolenko, U DeSilva and PM Muriana. Microbial profiling of biltong processing using culture-dependent and culture-independent microbiome analysis. Foods 2023; 12, 844.
LB Reller, MP Weinstein and CA Petti. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007; 44, 1108-14.
DM Hillis and JJ Bull. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993; 42, 182-92
KD Silva, RC Quisen, JD Goldbach, KBF Pepe and ANK Filho. Plant growth-promoting endophytic bacteria in peach palm seedlings. Pesquisa Agropecuária Bras. 2022; 57, e02962.
GT Dinkwar, K Thakur, S Bramhankar, T Pillai, S Isokar, T Ravali and V Kharat. Biochemical and physiological characterization of Bradyrhizobium japonicum. Int. J. Chem. Stud. 2020; 8, 1589-94.
M Sridevi and KV Mallaiah. Phosphate solubilization by Rhizobium strains. Indian J. Microbiol. 2009; 49, 98-102.
GK Kumar and MR Ram. Phosphate solubilizing rhizobia isolated from Vigna trilobata. Am. J. Microbiol. Res. 2014; 2, 105-9.
D Shokri and G Emtiazi. Indole-3-acetic acid (iaa) production in symbiotic and non-symbiotic nitrogen-fixing bacteria and its optimization by taguchi design. Curr. Microbiol. 2010; 61, 217-25.
S Ghosh and PS Basu. Production and metabolism of indole acetic acid in roots and root nodules of Phaseolus mungo. Microbiol. Res. 2006; 161, 362-6.
S Purwaningsih, D Agustiyani and S Antonius. Diversity, activity, and effectiveness of Rhizobium bacteria as plant growth promoting rhizobacteria (PGPR) isolated from Dieng, Central Java. Iranian J. Microbiol. 2021; 13, 130-6.
Downloads
Published
Issue
Section
License
Copyright (c) 2024 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






