First Full-Length Cloning of Human NaV1.7 in S. Cerevisiae-Based Novel D-Crypt^TM Platform for its High-Scale Production
DOI:
https://doi.org/10.48048/tis.2024.7232Keywords:
NaV1.7 Voltage-gated sodium channel, Fluorescence resonance energy transfer, Saccharomyces cerevisiae, Sodium channel blockersAbstract
NaV1.7, a voltage-gated sodium channel, induces chronic pain. Current research on the discovery of inhibitors against NaV1.7 is advancing with the hope of treating chronic pain conditions in patients suffering from various diseases. However, to characterize NaV1.7 and inhibitor interactions, a higher yield of expression is required to obtain sufficient protein. Molecular cloning of hNaV1.7 was done using the homologous recombination method in our novel S. cerevisiae-based D-cryptÔ platform. The platform was designed for the high-yield production of ‘difficult-to-express’ proteins (DTE-Ps) up to 500 L. The changes in the cell membrane potential of hNaV1.7 were determined using a fluorescence resonance energy transfer (FRET) assay with tetracaine (hNaV1.7 inhibitor). Expression analysis of hNaV1.7 showed 2 bands of size 250 and 280 kDa that were absent in untransfected yeast cells. Immunofluorescence images revealed the presence of hNaV1.7 on the membrane of hNaV1.7-expressing yeast cells. Tetracaine exhibited concentration-dependent inhibition of the FRET ratio, with the order of potency (IC50 = 0.46 µM) being approximately the same as previously reported, suggesting the functionality of the channel protein expressed in the D-cryptÔ platform. Cloning of NaV1.7 in the D-cryptÔ platform will help to scale up the production of channel proteins, which will ultimately help in the structural and functional characterization of the binding interactions between toxins and the NaV1.7 channel to identify more specific NaV1.7 inhibitors.
HIGHLIGHTS
- 7 is a voltage-gated sodium channel and induces chronic pain
- 7 inhibitors can advance the treatment of chronic pain associated with various diseases
- To characterize interaction of Nav1.7 with its inhibitors, higher yield of the NaV1.7 is required
- D-crypt is a platform designed for the high-yield production of ‘difficult-to-express’ proteins (DTE-Ps) up to 500 L
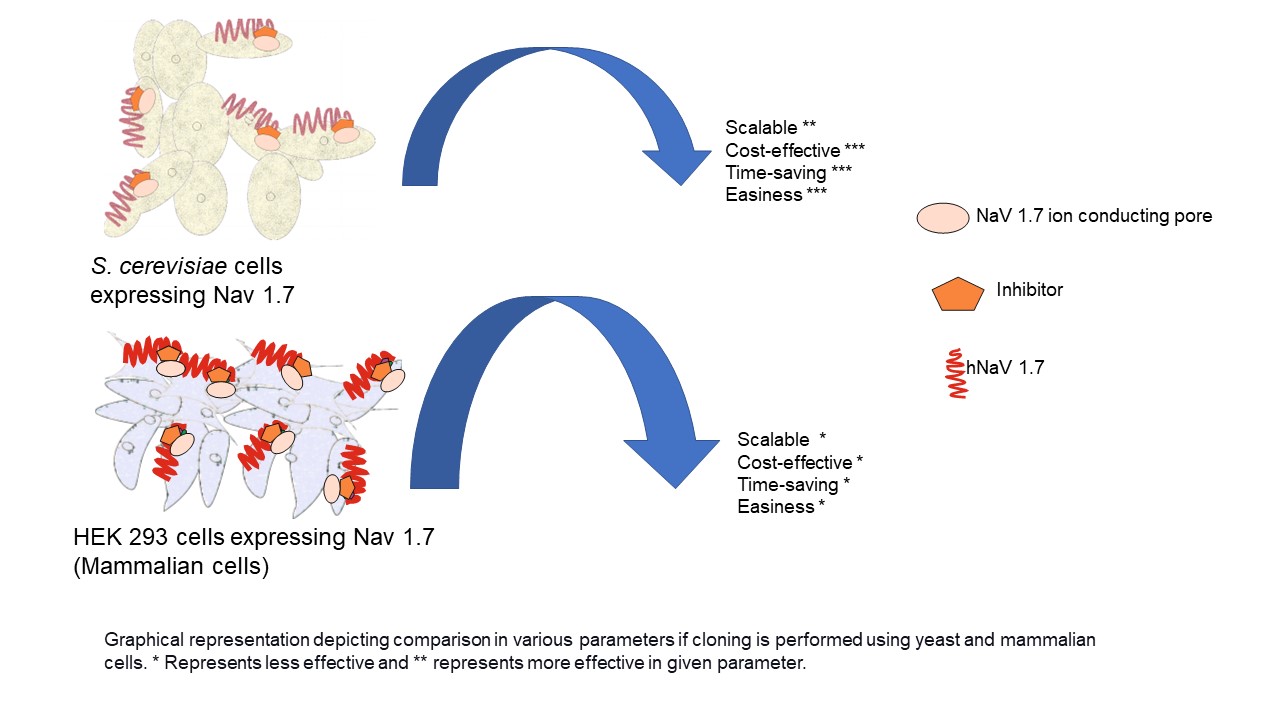
- Successful cloning of NaV1.7 in D-crypt platform suggest that the D-crypt platform is a suitable platform for expressing full-length functionally active hNaV1.7 channel at larger scale. It will further aid in the screening of NaV1.7 inhibitors
GRAPHICAL ABSTRACT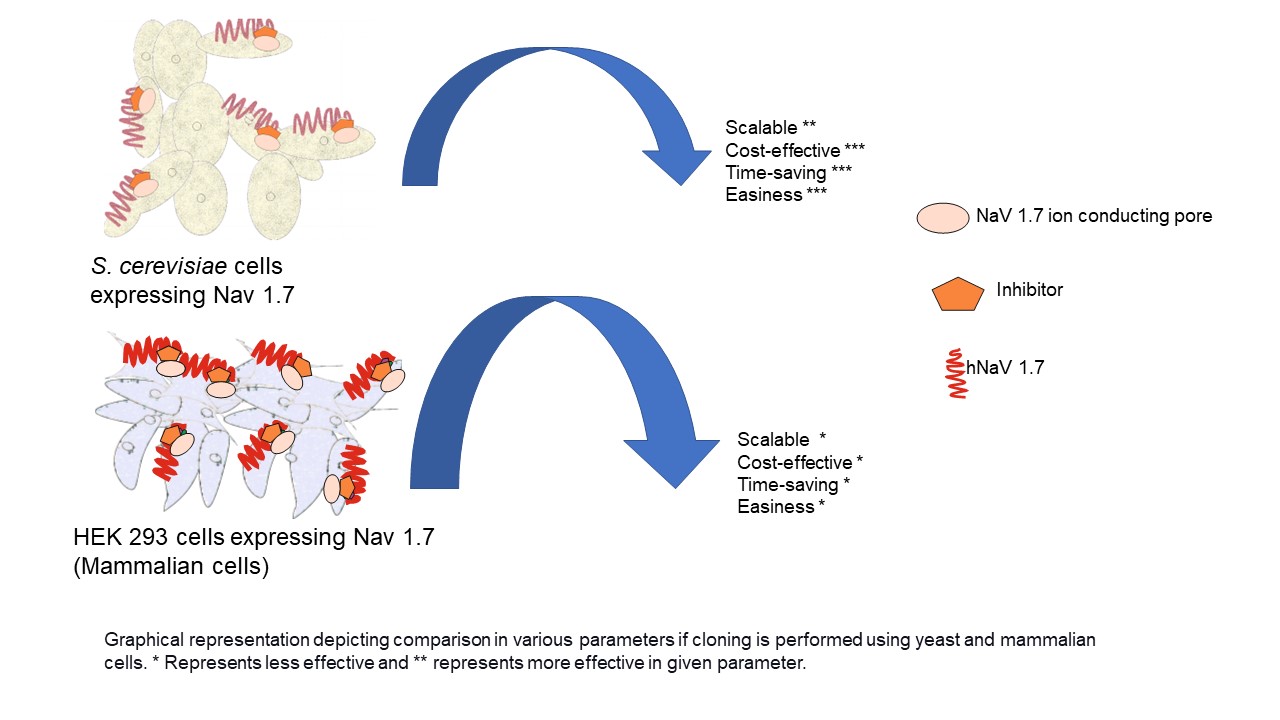
Downloads
Metrics
References
SD Dib-Hajj, TR Cummins, JA Black and SG Waxman. Sodium channels in normal and pathological pain. Annu. Rev. Neurosci. 2010; 33, 325-47.
JJ Cox, F Reimann, AK Nicholas, G Thornton, E Roberts, K Springell, G Karbani, H Jafri, J Mannan, Y Raashid, L Al-Gazali, H Hamamy, EM Valente, S Gorman, R Williams, DP McHale, JN Wood, FM Gribble and CG Woods. An SCN9A channelopathy causes congenital inability to experience pain. Nature 2006; 444, 894-8.
MD Baker and MA Nassar. Painful and painless mutations of SCN9A and SCN11A voltage-gated sodium channels. Pflügers Arch. 2020; 472, 865-80.
S Trivedi, A Pandit, G Ganguly and SK Das. Epidemiology of peripheral neuropathy: An Indian perspective. Ann. Indian Acad. Neurol. 2017; 20, 173-84.
JW Theile, BW Jarecki, AD Piekarz and TR Cummins. Nav1.7 mutations associated with paroxysmal extreme pain disorder, but not erythromelalgia, enhance Navbeta4 peptide-mediated resurgent sodium currents. J. Physiol. 2011; 589, 597-608.
G Sampoerno, J Sunariani and Kuntaman. Expression of NaV-1.7, TNF-alpha and HSP-70 in experimental flare-up post-extirpated dental pulp tissue through a neuroimmunological approach. Saudi Dent. J. 2020; 32, 206-12.
K Kingwell. Nav1.7 withholds its pain potential. Nat. Rev. Drug Discov. 2019; 18, 321-23.
J Gingras, S Smith, DJ Matson, D Johnson, K Nye, L Couture, E Feric, R Yin, BD Moyer, ML Peterson, JB Rottman, RJ Beiler, AB Malmberg and SI McDonough. Global Nav1.7 knockout mice recapitulate the phenotype of human congenital indifference to pain. PLos One 2014; 9, e105895.
S Hameed. Nav1.7 and Nav1.8: Role in the pathophysiology of pain. Mol. Pain 2019; 15, 1744806919858801.
ML Garcia and GJ Kaczorowski. Ion channels find a pathway for therapeutic success. Proc. Natl. Acad. Sci. Unit. States Am. 2016; 113, 5472-4.
S Trivedi, K Dekermendjian, R Julien, J Huang, L Per-Eric, J Krupp, R Kronqvist, O Larsson and R Bostwick. Cellular HTS assays for pharmacological characterization of Na(V)1.7 modulators. Assay Drug Dev. Tech. 2008; 6, 167-79.
S Mazumder, R Rastogi, A Undale, K Arora, NM Arora, B Pratim, D Kumar, A Joseph, B Mali, VB Arya, S Kalyanaraman, A Mukherjee, A Gupta, S Potdar, SS Roy, D Parashar, J Paliwal, SK Singh, A Naqvi, ..., PK Kundu. PRAK-03202: A triple antigen virus-like particle vaccine candidate against SARS CoV-2. Heliyon 2021; 7, e08124.
CJ Liu, BT Priest, RM Bugianesi, PM Dulski, JP Felix, IE Dick, RM Brochu, K Hans-Guenther, RE Middleton, GJ Kaczorowski, RS Slaughter, ML Garcia and MG Köhler. A high-capacity membrane potential FRET-based assay for NaV1.8 channels. Assay Drug Dev. Tech. 2006; 4, 37-48.
CJ Laedermann, N Syam, M Pertin, I Decosterd and H Abriel. β1- and β3- voltage-gated sodium channel subunits modulate cell surface expression and glycosylation of Nav1.7 in HEK293 cells. Front. Cell. Neurosci. 2013; 7, 137.
DH Feldman and C Lossin. The Nav channel bench series: Plasmid preparation. MethodsX 2014; 1, 6-11.
JM DeKeyser, CH Thompson and AL George. Cryptic prokaryotic promoters explain instability of recombinant neuronal sodium channels in bacteria. J. Biol. Chem. 2021; 296, 100298.
W Choi, H Ryu, A Fuwad, S Goh, C Zhou, J Shim, M Takagi, S Kwon, SM Kim and J Tae-Joon. Quantitative analysis of the membrane affinity of local anesthetics using a model cell membrane. Membranes 2021; 11, 579.
F Pomati, C Rossetti, D Calamari and BA Neilan. Effects of saxitoxin (STX) and veratridine on bacterial Na+ −K+ fluxes: A prokaryote-based STX bioassay. Appl. Environ. Microbiol. 2003; 69, 7371-6.
ER Benjamin, F Pruthi, S Olanrewaju, VI Ilyin, G Crumley, E Kutlina, KJ Valenzano and RM Woodward. State-dependent compound inhibition of Nav1.2 sodium channels using the FLIPR Vm dye: On-target and off-target effects of diverse pharmacological agents. J. Biomol. Screen. 2006; 11, 29-39.
RY Shimojo and WT Iwaoka. A rapid hemolysis assay for the detection of sodium channel-specific marine toxins. Toxicology 2000; 154, 1-7.
AM Smith, R Ammar, C Nislow and G Giaever. A survey of yeast genomic assays for drug and target discovery. Pharmacol. Therapeut. 2010; 127, 156-64.
H Shen, D Liu, K Wu, J Lei and N Yan. Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science 2019; 363, 1303-8.
Premas Biotech Private Limited, Available at: https://www.premasbiotech.com/biotalknology/casestudies, accessed April 2023.
T Chernov-Rogan, T Li, G Lu, H Verschoof, K Khakh, SW Jones, MH Beresini, C Liu, DF Ortwine, SJ McKerrall, DH Hackos, D Sutherlin, CJ Cohen and J Chen. Mechanism-specific assay design facilitates the discovery of Nav1.7-selective inhibitors. Proc Natl Acad Sci Unit. States Am. 2018; 115, E792-E801.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






