Production of Methanol Tolerant Cell-bound Lipase of Magnusiomyces spicifer SPB2 and Application as Whole-cell Biocatalyst in Transesterification Reaction
DOI:
https://doi.org/10.48048/tis.2023.6633Keywords:
Magnusiomyces spicifer SPB2, Hydrolysis activity, Cell-bound lipase activity, Transesterification activity, Taguchi orthogonal array method (L9), Whole-cell lipase, Fatty acid alkyl esterAbstract
Cell-bound lipase (CBL) is a potential biocatalyst for production of fatty acid alkyl ester (FAAE), which is feasible replacement for diesel fuel. This study aimed to improve both hydrolysis and transesterification activities of CBL from the yeast Magnusiomyces spicifer SPB2, which could be employed as whole-cell biocatalyst for transesterification reactions through optimizing the growth medium using Taguchi orthogonal array experimental method (L9). The optimized medium, resulting the highest CBL transesterification activity, consisted of soybean oil, peptone, Gum Arabic and an initial pH of 5. Interestingly, the fatty acid methyl ester (FAME) yield increased up to 33.28 times when compared with non-optimized medium. Moreover, cell morphology of M. spicifer SPB2 varied depending on the culture medium component. The single cell was more active to be used as the whole-cell biocatalyst than the pseudomycelial form for transesterification reaction. The highest FAME yield was 93.86 % when refined palm oil to methanol molar ratio of 1:8 was used as substrates. Whole-cell lipase from M. spicifer showed favorable activity to iso-butanol as an acyl accepter, resulting in fatty acid butyl ester (FABE) yield of 86.80 % at 1:3 molar ratio of palm oil to iso-butanol. The FAME and FABE yields dramatically decreased from 93.86 and 86.80 % to 57.02 and 4.48 %, respectively after the reuse of the enzyme to the second batch of reaction. These results indicated that the whole-cell biocatalyst developed from M. spicifer was an efficient and economical approach to improve enzyme activity and stability in the transesterification reaction.
HIGHLIGHTS
- Methanol tolerant cell-bound lipase and extracellular lipase of spicifer SPB2 were produced
- The whole-cell biocatalyst of spicifer SPB2 was developed for FAME production
- The optimized medium was developed to prepare whole-cell lipases from spicifer SPB2
- Gum Arabic was the most influencing factor for synthetic activity of whole-cell CBL from yeast
- Single cell morphology exhibited higher transesterification activity than pseudomycelial form
GRAPHICAL ABSTRACT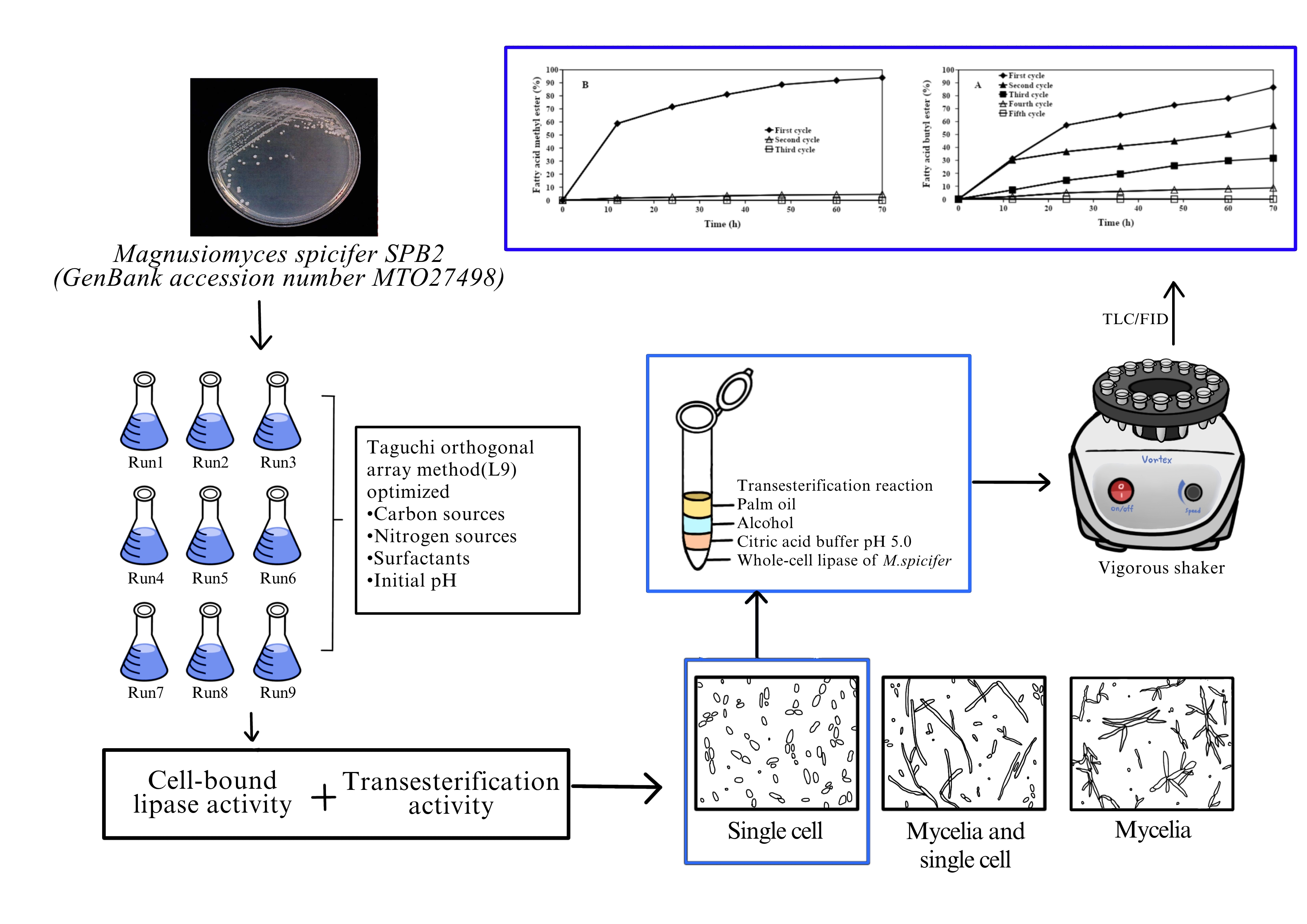
Downloads
References
O Ilesanmi, AE Adekunle, JA Omolaiye, EM Olorode and AL Ogunkanmi. Isolation, optimization and molecular characterization of lipase producing bacteria from contaminated soil. Sci. Afr. 2020; 8, e00279.
S Verma, GK Meghwanshi and R Kumar. Current perspectives for microbial lipases from extremophiles and metagenomics. Biochimie 2021; 182, 23-36.
S Fatima, A Faryad, A Ataa, FA Joyia and A Parvaiz. Microbial lipase production: A deep insight into the recent advances of lipase production and purification techniques. Biotechnol. Appl. Biochem. 2020; 68, 445-58.
C Bhan and J Singh. Role of microbial lipases in transesterifcation process for biodiesel production. J. Environ. Sustain. 2020; 3, 257-66.
F Toldrá-Reig, L Mora and F Toldrá. Developments in the use of lipase transesterification for biodiesel production from animal fat waste. Appl. Sci. 2020; 10, 5085.
A Haq, S Adeel, A Khan, Q Rana, MAN Khan, M Rafiq, M Ishfaq, S Khan, AA Shah, F Hasan, S Ahmed and M Badshah. Screening of lipase-producing bacteria and optimization of lipase-mediated biodiesel production from Jatropha curcas seed oil using whole cell approach. Bioenerg. Res. 2020; 13, 1280-96.
N Rachmadona, E Quayson, J Amoah, DA Alfaro-Sayes, S Hama, M Aznury, A Kondo and C Ogino. Utilizing palm oil mill effluent (POME) for the immobilization of Aspergillus oryzae whole-cell lipase strains for biodiesel synthesis. Biofuels Bioprod. Bioref. 2021; 15, 804-14.
A Arumugam and V Ponnusami. Biodiesel production from Calophyllum inophyllum oil using lipase producing Rhizopus oryzae cells immobilized within reticulated foams. Renew. Energ. 2014; 64, 276-82.
SS Mohamed, HM Ahmed, MA El-Bendary, ME Moharam and HA Amin. Response surface methodology for optimization of Rhizopus stolonifer 1aNRC11 mutant F whole-cell lipase production as a biocatalyst for methanolysis of waste frying oil. Biocatal. Biotransformation 2021; 39, 232-40.
P Srimhan, K Kongnum, S Taweerodjanakarn and T Hongpattarakere. Selection of lipase producing yeasts for methanol-tolerant biocatalyst as whole cell application for palm-oil transesterification. Enzyme Microb. Tech. 2011; 48, 293-98.
MD Pramitasari and M Ilmi. Optimization of medium for lipase production from Zygosaccharomyces mellis SG1.2 using statistical experiment design. Microbiol. Biotechnol. Lett. 2021; 49, 337-45.
D Wang, Y Xu and T Shan. Effects of oils and oil-related substrates on the synthetic activity of membrane-bound lipase from Rhizopus chinensis and optimization of the lipase fermentation media. Biochem. Eng. J. 2008; 41, 30-7.
M Lopes, SM Miranda, JM Alves, AS Pereira and I Belo. Waste cooking as feedstock for lipase and lipid-rich biomass production. Eur. J. Lipid Sci. Tech. 2019; 121, 1800188.
JS Koh, T Kodama and Y Minoda. Screening of yeasts and cultural conditions for cell production from palm oil. Agric. Biol. Chem. 1983; 47, 1207-12.
A Nuylert and T Hongpattarakere. Improvement of cell-bound lipase from Rhodotorula mucilaginosa P11I89 for use as a methanol-tolerant, whole-cell biocatalyst for production of palm-oil biodiesel. Ann. Microbiol. 2013; 63, 929-39.
T Leevijit, C Tongurai, G Prateepchaikul and W Wisutmethangoon. Performance test of a 6-stage continuous reactor for palm methyl ester production. Bioresour. Tech. 2008; 99, 214-21.
MR Green and J Sambrook. Isolation of high-molecular-weight DNA using organic solvents. Cold Spring Harb. Protoc. 2017; 4, 356-60.
TJ White, T Bruns, S Lee and J Taylor. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. Academic press, San Diego, 1990, p. 315.
T Tan, M Zhang, J Xu and J Zhang. Optimization of culture conditions and properties of lipase from Penicillium camembertii Thom PG-3. Process Biochem. 2004; 39, 1495-502.
RR Maldonado, JFM Burkert, MA Mazutti, F Maugeri and MI Rodrigues. Evaluation of lipase production by Geotrichum candidum in shaken flasks and bench-scale stirred bioreactor using different impellers. Biocatal. Agr. Biotechnol. 2012; 1, 147-51.
AF de Almeida, SM Taulk-Tornisielo and EC Carmona. Influence of carbon and nitrogen sources on lipase production by a newly isolated Candida viswanathii strain. Ann. Microbiol. 2013, 63, 1225-34.
AR Byreddy, NM Rao, CJ Barrow and M Puri. Tween 80 influences the production of intracellular lipase by Schizochytrium S31 in a stirred tank reactor. Process Biochem. 2017; 53, 30-5.
S Moradi, SH Razavi and SM Mousavi. Isolation of lipase producing bacteria from olive extract to improve lipase production using a submerge fermentation technique. J. Food Bioprocess Eng. 2018; 1, 1-6.
N Cihangir and E Sarikaya. Investigation of lipase production by a new of Aspergillus sp. World J. Microbiol. Biotechnol. 2004; 20, 193-7.
A Najjar, EA Hassan, N Zabermawi, SH Saber, LH Bajrai, MS Almuhayawi, TS Abujamel, SB Almasaudi, LE Azhar, M Moulay and S Harakeh. Optimizing the catalytic activities of methanol and thermotolerant Kocuria fava lipases for biodiesel production from cooking oil wastes. Sci. Rep. 2021; 11, 13659.
K Ban, M Kaieda, T Matsumoto, A Kondo and H Fukuda. Whole cell biocatalyst for biodiesel fuel production utilizing Rhizopus oryzae cells immobilized within biomass support particles. Biochem. Eng. J. 2001; 8, 39-43.
GI de Becze. A microbiological process report; yeasts. I. Morphology. Appl. Microbiol. 1956; 4, 1-12.
AS Kokoreva, EP Isakova, VM Tereshina, OI Kiein, NN Gessler and YI Deryyabina. The effect of different substrates on the morphological features and polyols production of Endomyces magnusii yeast during long-lasting cultivation. Microorganisms 2022; 10, 1709.
M Kieliszek, S Błażejak, A Bzducha-Wróbel and AM Kot. Effect of selenium on lipid and amino acid metabolism in yeast cells. Biol. Trace Elem. Res. 2019; 187, 316-27.
LC Almeida, MS Barbosa, FA de Jesus, RM Santos, AT Fricks, LS Freitas, MM Pereira, ȦS Lima and CMF Soares. Enzymatic transesterification of coconut oil by using immobilized lipase on biochar: An experimental and molecular docking study. Biotechnol. Appl. Biochem. 2021; 68, 801-8.
J Liu, G Chen, B Yan, W Yi and J Yao. Biodiesel production in a magnetically fluidized bed reactor using whole-cell biocatalysts immobilized within ferroferric oxide-polyvinyl alcohol composite beads. Bioresour. Tech. 2022; 355, 127253.
OJ Chioke, CN Ogbonna, C Onwusi and JC Ogbonna. Lipase in biodiesel production. Afr. J. Biochem. Res. 2018; 12, 73-85.
A Salis, M Pinna, M Monduzzi and V Solinas. Biodiesel production from triolein and short chain alcohols through biocatalysis. J. Biotechnol. 2005; 119, 291-9.
A Arumugam and V Ponnusami. Production of biodiesel by enzymatic transesterification of waste sardine oil and evaluation of its engine performance. Renew Energ. 2017; 3, e00486.
Y Shimada, Y Watanabe, A Sugihara and Y Tominaga. Enzymatic alcoholysis for biodiesel fuel production and application of the reaction to oil processing. J. Mol. Catal. B Enzym. 2002; 17, 133-42.
Y Liu, L Huang, D Zheng, Y Fu, M Shan, Z Xu, J Ma and F Lu. Development of a Pichia pastoris whole-cell biocatalyst with overexpression of mutant lipase I PCLG47I from Penicillium cyclopium for biodiesel production. RSC Adv. 2018; 8, 26161-8.
MM Soumanou and UT Bornscheuer. Lipase-catalyzed alcoholysis of vegetable oils. Eur. J. Lipid Sci. Tech. 2003; 105, 656-60.
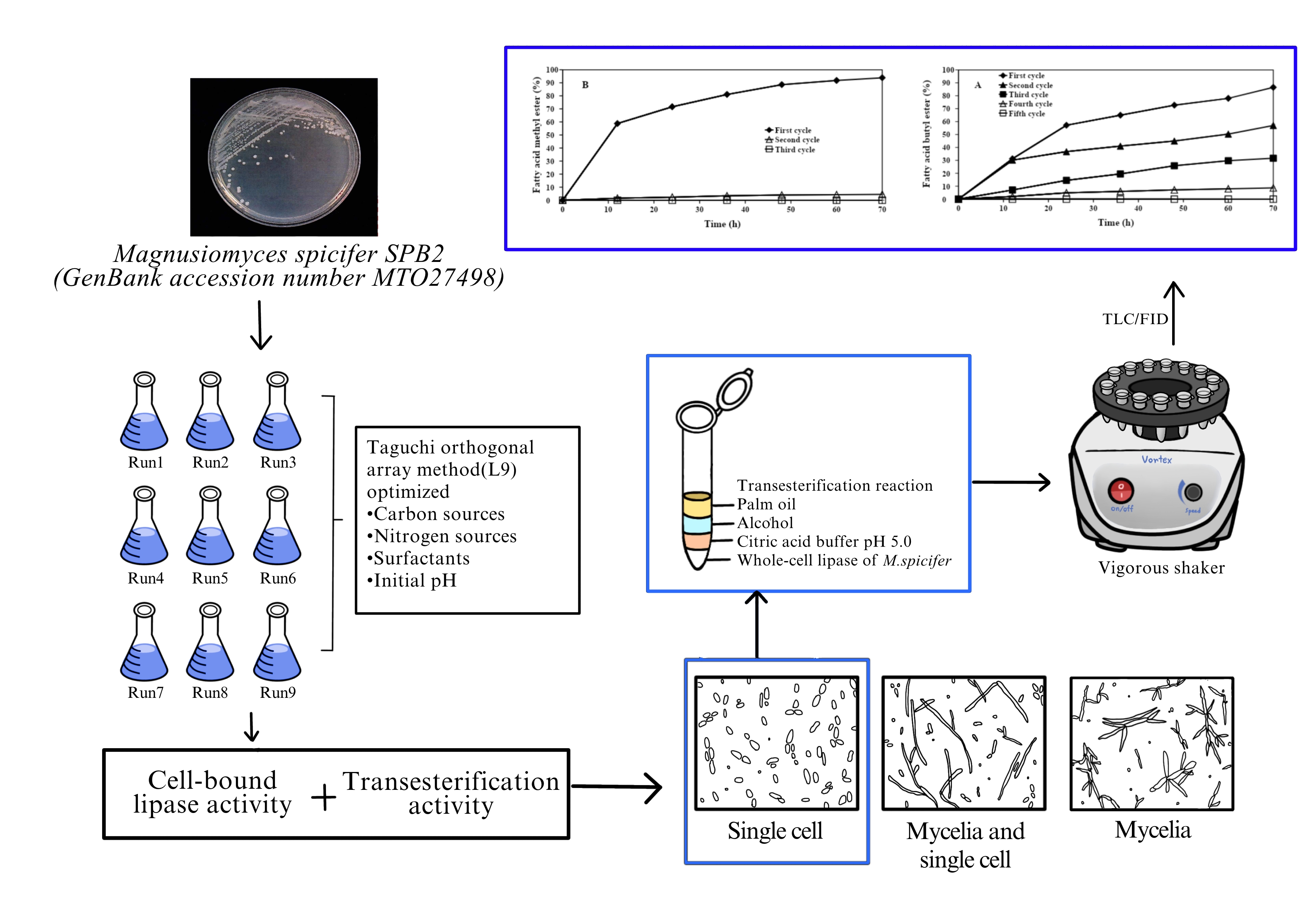
Downloads
Published
Issue
Section
License
Copyright (c) 2023 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






