Novel SNP Markers and Their Application in Low-Cyanide Cassava (Manihot esculenta crantz) Breeding Program
DOI:
https://doi.org/10.48048/tis.2023.6456Keywords:
SNP detection, Tetra-primer ARMS-PCR, Cyanogenic glycoside, Hydrocyanic acid, Molecular breedingAbstract
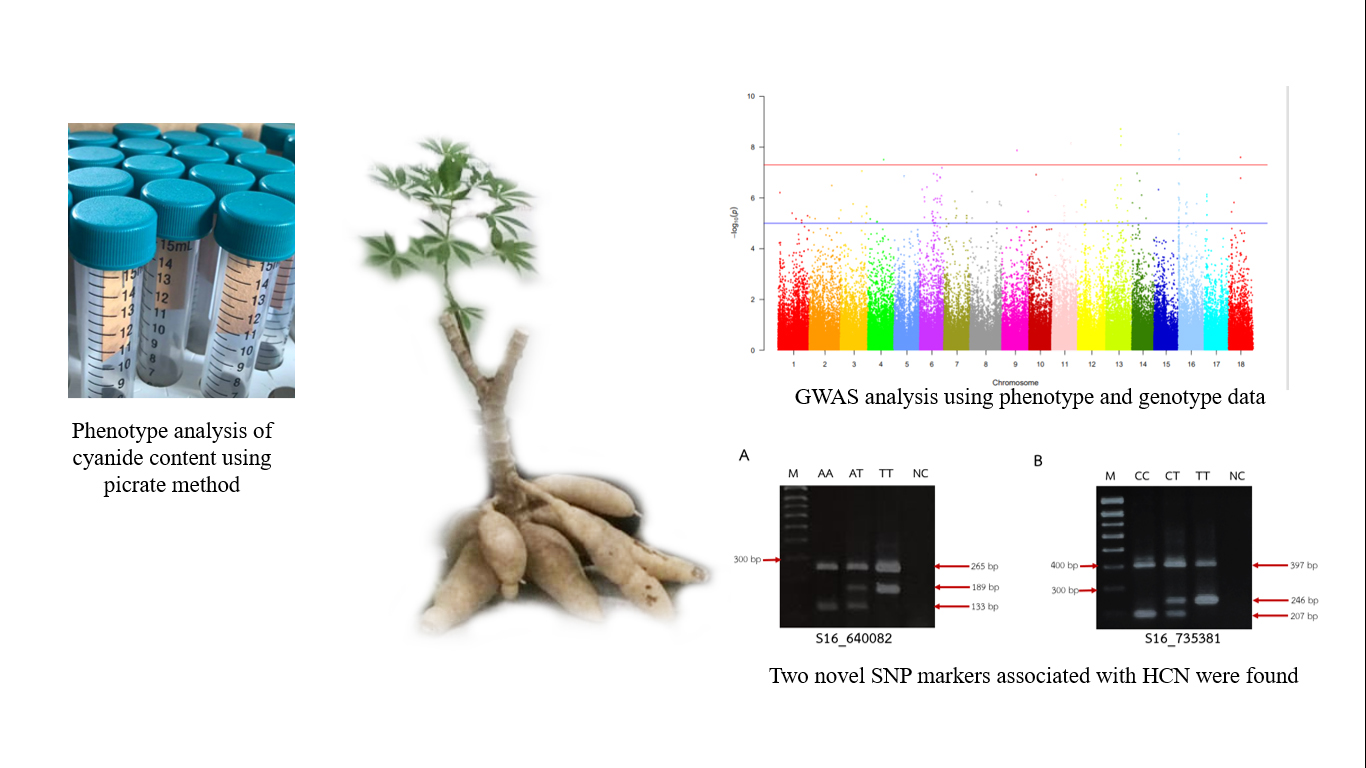
The major flaw of cassava is its indigenous cyanide. Traditional breeding program for diminishing cyanide in cassava take a long waiting time for harvesting cassava tubers to assess cyanide content. Genetic markers are useful for selection of good agricultural traits, which also applied in low-cyanide cassava breeding programs by detecting gene associate with cyanide content in cassava. Therefore, the objective of this study was to develop SNP markers to detect low cyanide trait in cassava. Genotype analysis was carried out using Genotyping by Sequencing (GBS) technology assembling with phenotype analysis. Then, the SNP markers associated with cyanide content were found. Afterward, the primers of SNP marker associated with cyanide content 2 positions were developed using tetra-primer ARMS-PCR techniques. S16_640082 and S16_735381 SNP markers on chromosome 16 showed potential ability of the selection of cassava varieties with cyanide content less than 250 mg HCN/kg fresh weight. The selection accuracy of S16_640082 and S16_735381 SNP markers was 73.33 and 76.64 %, respectively. These markers could be used in marker assisted selection (MAS) in cassava seedling. It substantially reduces cassava breeding programs cost and time.
HIGHLIGHTS
- The major flaw of cassava is its indigenous cyanide. Up to our knowledge, there is rarely report on SNP marker related to cyanide content in Thai cassava population
- By performing genome wide association mapping (GWAS) to characterize the genome position related to HCN, 2 novel SNP markers associated with cyanide content were found
- The primers to detect the SNP markers were developed using tetra-primer ARMS-PCR techniques
- The markers showed ability of the selection of cassava varieties with cyanide content less than 250 mg HCN/kg fresh weight
GRAPHICAL ABSTRACT
Downloads
Metrics
References
FAOSTAT, Available at: https://www.fao.org/faostat, accessed June 2020.
D Tshala-Katumbay, N Mumba, L Okitundu, K Kazadi, M Banea, T Tylleskär, M Boivin and JJ Muyembe-Tamfum. Cassava food toxins, konzo disease, and neurodegeneration in sub-Sahara Africans. Neurology 2013; 80, 949-51.
E Kashala-Abotnes, D Okitundu, D Mumba, MJ Boivin, T Tylleskär and D Tshala-Katumbay. Konzo: A distinct neurological disease associated with food (cassava) cyanogenic poisoning.
Brain Res. Bull. 2019; 145, 87-91.
E Rivadeneyra-Domínguez and JF Rodríguez-Landa. Preclinical and clinical research on the toxic and neurological effects of cassava (Manihot esculenta Crantz) consumption. Metab. Brain Dis. 2020; 35, 65-74.
H Ceballos, JC Pérez, BO Joaqui, JI Lenis, N Morante, F Calle, L Pino and CH Hershey. Cassava breeding I: The value of breeding value. Front. Plant Sci. 2016; 7, 1227.
E Balyejusa Kizito, AC Rönnberg-Wästljung, T Egwang, U Gullberg, M Fregene and A Westerbergh. Quantitative trait loci controlling cyanogenic glucoside and dry matter content in cassava (Manihot esculenta Crantz) roots. Hereditas 2007; 144, 129-36.
S Whankaew, S Poopear, S Kanjanawattanawong, S Tangphatsornruang, O Boonseng, DA Lightfoot and K Triwitayakorn. A genome scan for quantitative trait loci affecting cyanogenic potential of cassava root in an outbred population. BMC Genom. 2011; 12, 266.
MM Rana, T Takamatsu, M Baslam, K Kaneko, K Itoh, N Harada, T Sugiyama, T Ohnishi, T Kinoshita, H Takagi and T Mitsui. Salt tolerance improvement in rice through efficient SNP marker-assisted selection coupled with speed-breeding. Int. J. Mol. Sci. 2019; 20, 2585.
S Ravi, M Hassani, B Heidari, s Deb, E Orsini, J Li, CM Richards, LW Panella, S Srinivasan, G Campagna, G Concheri, A Squartini and P Stevanato. Development of an SNP assay for marker-assisted selection of soil-borne Rhizoctonia solani AG-2-2-IIIB resistance in sugar beet. Biology 2021; 11, 49.
Z Chen, D Tang, J Ni, P Li, L Wang, J Zhou, C Li, H Lan, L Li and J Liu. Development of genic KASP SNP markers from RNA-Seq data for map-based cloning and marker-assisted selection in maize. BMC Plant Biol. 2021; 21, 157.
RS Kawuki, M Ferguson, M Labuschagne, L Herselman and DJ Kim. Identification, characterisation and application of single nucleotide polymorphisms for diversity assessment in cassava (Manihot esculentaCrantz). Mol. Breed. 2009; 23, 669-84.
AC Ogbonna, LRBD Andrade, IY Rabbi, LA Mueller, EJD Oliveira and GJ Bauchet. Genetic architecture and gene mapping of cyanide in cassava (Manihot esculenta Crantz.). bioRxiv 2020. https://doi.org/10.1101/2020.06.19.159160
AD Ige, B Olasanmi, EGN Mbanjo, IS Kayondo, EY Parkes, P Kulakow, C Egesi, GJ Bauchet, E Ng, LAB Lopez-Lavalle, H Ceballos and IY Rabbi. Conversion and validation of uniplex SNP markers for selection of resistance to cassava mosaic disease in cassava breeding programs. Agronomy 2021; 11, 420.
MG Bradbury, SV Egan and JH Bradbury. Determination of all forms of cyanogens in cassava roots and cassava products using picrate paper kits. J. Sci. Food Agr. 1999; 79, 593-601.
MW Blair, MA Fregene, SE Beebe and H Ceballos. Marker-assisted selection in common beans and cassava. In: EP Guimaraes, J Ruane, BD Scherf, A Sonnino and JD Dargie (Eds.). Marker-assisted selection: Current status and future perspectives in crops, livestock, forestry and fish. Organización de las Naciones Unidas para la Agricultura y la Alimentación, Rome, Italy, 2007, p. 81-115.
Y He, C Zhou, M Huang, C Tang, X Liu, Y Yue, Q Diao, Z Zheng and D Liu. Glyoxalase system: A systematic review of its biological activity, related-diseases, screening methods and small molecule regulators. Biomed. Pharmacother. 2020; 131, 110663.
OA Gyamfi, N Bortey-Sam, SB Mahon, M Brenner, GA Rockwood and BA Logue. Metabolism of cyanide by glutathione to produce the novel cyanide metabolite 2-aminothiazoline-4-oxoaminoethanoic acid. Chem. Res. Toxicol. 2019; 32, 718-26.
G Brychkova, A Kurmanbayeva, A Bekturova, I Khozin, D Standing, D Yarmolinsky and M Sagi. Determination of enzymes associated with sulfite toxicity in plants: Kinetic Assays for SO, APR, SiR, and In-Gel SiR Activity. Meth. Mol. Biol. 2017; 1631, 229-51.
EU Maduh, EW Nealley, H Song, PC Wang and SI Baskin. A protein kinase C inhibitor attenuates cyanide toxicity in vivo. Toxicol. 1995; 100, 129-37.
S Zhang. Activation of plant stress-responsive MAP kinases induces cyanide - its role in reactive oxygen species generation and hypersensitive cell death, Available at: https://grantome.com/ grant/NSF/IOS-0743957, accessed May 2022.
B Darbani, MS Motawia, CE Olsen, HH Nour-Eldin, BL Møller and F Rook. the biosynthetic gene cluster for the cyanogenic glucoside dhurrin in sorghum bicolor contains its co-expressed vacuolar MATE transporter. Sci. Rep. 2016; 6, 37079.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






