In silico Analysis and Characterization of a Putative Aspartic Proteinase Inhibitor, IA3 from Lachancea kluyveri
DOI:
https://doi.org/10.48048/tis.2023.6377Keywords:
Bioinformatics, BLAST, QSAR, Aspartic proteinase, IA3Abstract
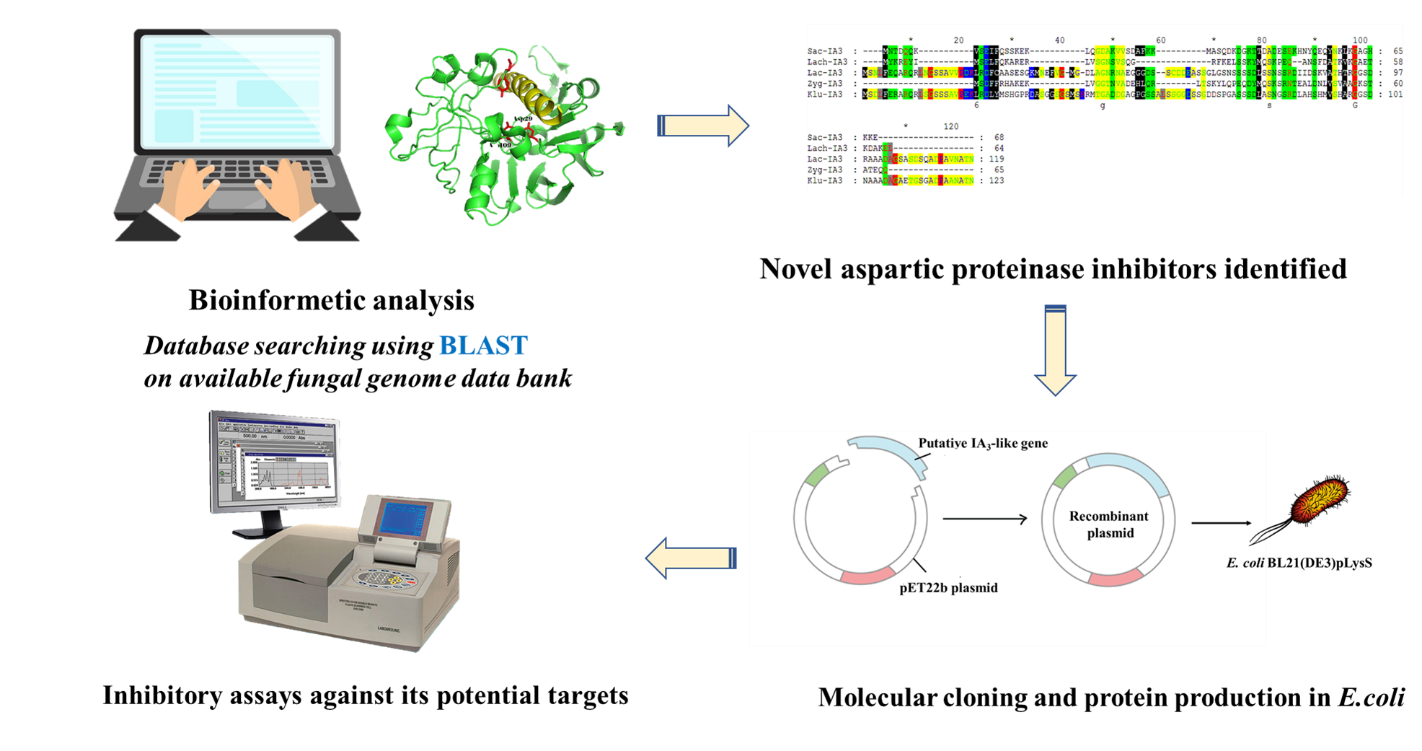
Aspartic proteinases play important role in various physiological and biological processes. Understanding catalytic mechanisms of these enzymes could lead to crucial medical and biological applications. Two types of aspartic proteinase inhibitors have been identified i.e. small molecule inhibitor and naturally occurring peptides. Most of aspartic proteinases are highly susceptible to inhibition by a series of small, non-proteinaceous natural products; pepstatin. However, only limited number of naturally-occurring polypeptide inhibitors of aspartic proteinases has so far been discovered. One among these inhibitors is Saccharomyces cerevisiae IA3. The S. cerevisiae Proteinase A (ScPrA) is solely and potently inhibited by this small polypeptide at sub-nanomolar level. It was proven that, not only IA3 has no detectable effect against a wide range of aspartic proteinases, but it was also shown to be cleaved as a substrate of these non-target enzymes. Bioinformatics analysis of the IA3 structure has been undertaken in this study. Database searching for sequence homology from available fungal genome data bank using the Basic Local Alignment Search Tool (BLAST) revealed that 4 structurally related, IA3-like proteins have successfully been identified. The amino acid sequence of IA3-like proteins from Lachancea kluyveri share highest degree of similarity toward wild-type IA3. The Lk-IA3-like gene was synthesized using a PCR-based gene synthesis method. Protein expression in E. coli, protein purification and characterization of Lk-IA3-like by enzyme kinetic assays were performed. The results indicated that Lk-IA3-like protein inhibits ScPrA at the Ki value of 190 ± 0.11 nM and possesses no inhibitory effect toward AfPrA (Aspergillus fumigatus proteinase A) or HuCatD (Human cathepsin D).
HIGHLIGHTS
- Aspartic proteinases enzyme family is crucially involved in various physiological and biological processes including in the pathogenesis of numerous diseases which make them a possible target for drug design
- Inhibitors of aspartic proteinases are not only applied for human disease treatment but also extended to plant and other economically important animals
- Aspartic proteinase from cerevisiae (Proteinase A) is solely and potently inhibited by a small peptideinhibitor, IA3
- Bioinformatics analysis of the naturally occurring aspartic proteinases inhibitor, IA3 structure has been undertaken in this research and putative IA3-like proteins have successfully been identified
- IA3-like proteins from Lachancea kluyveri were produced in coli and enzyme inhibition assays revealed that Lk-IA3-like protein inhibits ScPrA at the Ki value of 190 ± 0.11 nM
GRAPHICAL ABSTRACT 
Downloads
References
JM Berg, JL Tymoczko and L Stryer. Biochemistry. W.H. Freeman, Basingstoke, 2012.
AMD Castro, AFD Santos, V Kachrimanidou, AA Koutinas, DMG Freire. Solid-state fermentation for the production of proteases and amylases and their application in nutrient medium production. In: A Pandey, C Larroche and CR Soccol (Eds.). Current developments in biotechnology and bioengineering, Elsevier, Cambridge, 2018, p. 185-210.
ND Rawlings. Peptidase inhibitors in the MEROPS database. Biochimie 2010; 92, 1463-83.
ND Rawlings, AJ Barrett, PD Thomas, X Huang, A Bateman and RD Finn. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018; 46, D624-D632.
AR Khan amd MN James. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998; 7, 815-36.
BM Dunn. Aspartic proteases. In: WJ Lennarz and MD Lane (Eds.). Encyclopedia of biological chemistry. 2nd ed. Academic Press, Waltham, 2013, p. 137-40.
ND Rawlings and AJ Barrett. Introduction: Aspartic and glutamic peptidases and their clans. In: ND Rawlings and G Salvesen (Eds.). Handbook of proteolytic enzymes. 3rd ed. Academic Press, Cambridge, 2013, p. 3-19.
A Wlodawer, A Gustchina and MNG James. Catalytic pathways of aspartic peptidases. In: ND Rawlings and G Salvesen (Eds.). Handbook of proteolytic enzymes 3rd ed. Academic Press, Cambridge, 2013, p. 3-19.
JB Cooper. Aspartic proteinases in disease: A structural perspective. Curr. Drug Targets 2002; 3, 155-73.
J Kay. Aspartic proteinases and their inhibitors. Biochem. Soc. Trans. 1985; 13, 1027-9.
MJ Valler, J Kay, T Aoyagi and BM Dunn. The interaction of aspartic proteinases with naturally-occurring inhibitors from actinomycetes and Ascaris lumbricoides. J. Enzym. Inhib. 1985; 1, 77-82.
LH Phylip, WE Lees, BG Brownsey, D Bur, BM Dunn, JR Winther, A Gustchina, M Li, T Copeland, A Wlodawer and J Kay. The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae. J. Biol. Chem. 2001; 276, 2023-30.
S Takahashi, K Takahashi, T Kaneko, H Ogasawara, S Shindo, K Saito and Y Kawamura. Identification of functionally important cysteine residues of the human renin-binding protein as the enzyme N-acetyl-D-glucosamine 2-epimerase. J. Biochem. 2001; 129, 529-35.
S Takahashi, H Ogasawara, K Takahashi, K Hori, K Saito and K Mori. Identification of a domain conferring nucleotide binding to the N-acetyl-d-glucosamine 2-epimerase (Renin binding protein). J. Biochem. 2002; 131, 605-10.
G Dubin. Proteinaceous cysteine protease inhibitors. Cell Mol. Life Sci. 2005; 62, 653-69.
K Galesa, B Strukelj, S Bavec, V Turk and B Lenarcic. Cloning and expression of functional equistatin. Biol. Chem. 2000; 381, 85-8.
B Strukelj, B Lenarcic, K Gruden, J Pungercar, B Rogelj, V Turk, D Bosch and MA Jongsma. Equistatin, a protease inhibitor from the sea anemone Actinia equina, is composed of three structural and functional domains. Biochem. Biophys. Res. Commun. 2000; 269, 732-6.
B Lenarcic and V Turk. Thyroglobulin type-1 domains in equistatin inhibit both papain-like cysteine proteinases and cathepsin D. J. Biol. Chem. 1999; 274, 563-6.
B Lenarcic, A Ritonja, B Strukelj, B Turk and V Turk. Equistatin, a new inhibitor of cysteine proteinases from Actinia equina, is structurally related to thyroglobulin type-1 domain. J. Biol. Chem. 1997; 272, 13899-903.
T Kageyama. Molecular cloning, expression and characterization of an Ascaris inhibitor for pepsin and cathepsin E. Eur. J. Biochem. 1998; 253, 804-9.
T Kageyama. Pepsinogens, progastricsins, and prochymosins: Structure, function, evolution, and development. Cell Mol. Life Sci. 2002; 59, 288-306.
K Girdwood and C Berry. The disulphide bond arrangement in the major pepsin inhibitor PI-3 of Ascaris suum. FEBS Lett. 2000; 474, 253-4.
K Ng, JFW Petersen, M Cherney, C Garen, J Zalatoris, C Rao-Naik, B Dunn, M Martzen, R Peanasky and M James. Structural basis for the inhibition of porcine pepsin by Ascaris pepsin inhibitor-3. Nat. Struct. Biol. 2000; 7, 653-7.
T Dreyer, MJ Valler, J Kay, P Charlton and BM Dunn. The selectivity of action of the aspartic-proteinase inhibitor IA3 from yeast (Saccharomyces cerevisiae). Biochem. J. 1985; 231, 777-9.
M Li, LH Phylip, WE Lees, JR Winther, BM Dunn, A Wlodawer, J Kay and A Gustchina. The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix. Nat. Struct. Biol. 2000; 7, 113-7.
T Saheki, Y Matsuda and H Holzer. Urification and characterization of macromolecular inhibitors of proteinase A from yeast. Eur. J. Biochem. 1974; 47, 325-32.
K Biedermann, U Montali, B Martin, I Svendsen and M Ottesen. The amino acid sequence of proteinase a inhibitor 3 from baker’s yeast. Carlsberg Res. Commun. 1980; 45, 225.
P Schu and DH Wolf. The proteinase yscA-inhibitor, IA3, gene. Studies of cytoplasmic proteinase inhibitor deficiency on yeast physiology. FEBS Lett. 1991; 283, 78-84.
SF Altschul, W Gish, W Miller, EW Myers and DJ Lipman. Basic local alignment search tool. J. Mol. Biol. 1990; 215, 403-10.
JD Thompson, TJ Gibson, F Plewniak, F Jeanmougin and DG Higgins. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997; 25, 4876-82.
P Stothard. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000; 28, 1102.
C Withers-Martinez, EP Carpenter, F Hackett, B Ely, M Sajid, M Grainger and MJ Blackman. PCR-based gene synthesis as an efficient approach for expression of the A+T-rich malaria genome. Protein Eng. 1999; 12, 1113-20.
MM Bradford. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976; 72, 248-54.
TJ Winterburn, DM Wyatt, LH Phylip, C Berry, D Bur and J Kay. Adaptation of the behaviour of an aspartic proteinase inhibitor by relocation of a lysine residue by one helical turn. Biol. Chem. 2006; 387, 1139-42.
TJ Winterburn, DM Wyatt, LH Phylip, D Bur, RJ Harrison, C Berry and J Kay. Key features determining the specificity of aspartic proteinase inhibition by the helix-forming IA3 polypeptide. J. Biol. Chem. 2007; 282, 6508-16.
DM Kupfer, SD Drabenstot, KL Buchanan, H Lai, H Zhu, DW Dyer, BA Roe and JW Murphy. Introns and splicing elements of five diverse fungi. Eukaryot Cell 2004; 3, 1088-100.
Downloads
Published
Issue
Section
License
Copyright (c) 2023 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






