A Label-Free Marker Based Breast Cancer Detection using Hybrid Deep Learning Models and Raman Spectroscopy
DOI:
https://doi.org/10.48048/tis.2023.6299Keywords:
Breast cancer detection, Hybrid deep learning, Spectral data augmentation, 1Dimensional-CNN-GRUAbstract
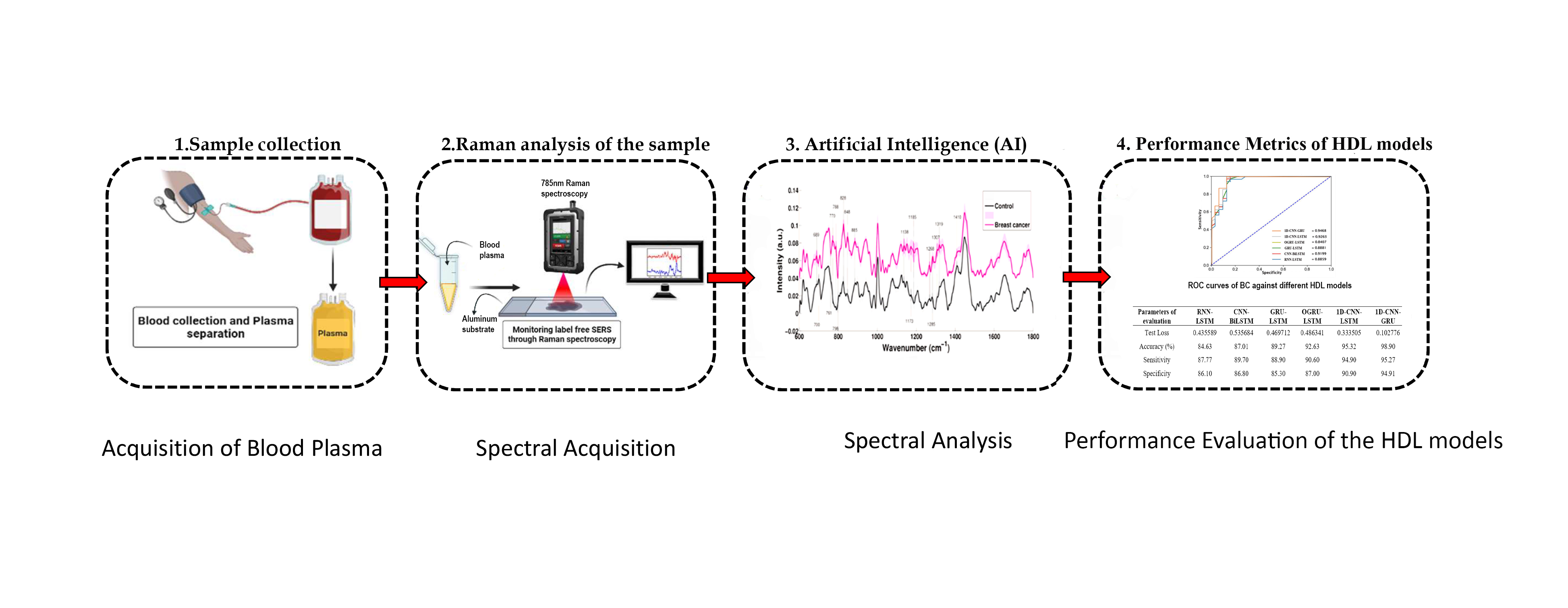
Breast Cancer (BC) is a serious menace to women’s health around the world. Early BC identification has been critically important for diagnosing protocol. Several classification methods for breast cancer were examined recently with various techniques, and Raman spectroscopy (RS) has become an effective approach for the identification of responsible metabolites. Moreover, the rapid and accurate classification of BC using RS necessitates active engagement in processing and analyzing Raman spectral data. This work aims to develop an efficient Hybrid Deep Learning (HDL) neural network model to differentiate breast cancer blood plasma from control samples and the spectral features obtained are used as spectral cancer markers for the detection of breast cancer. To find the optimum performing HDL model, several other HDL models were implemented to perform the binary classification of the Raman spectral signal. A total of 62199 Raman spectra generated from 26 blood plasma samples are evaluated in this study. Mainly 6 HDL methods, 1D-CNN-GRU, CNN-BiLSTM-AT, 1D-CNN-LSTM, GRU-LSTM, RNN-LSTM, and OGRU-LSTM are modeled to evaluate the performance of hybrid models to identify 2 classes of Raman spectral data. Comparative classification results show that the stacked 1D-CNN-GRU model outperforms well for breast cancer detection using the Raman spectral dataset than other prominent HDL architectures. The stacked 1D-CNN-GRUclassifier model achieved the highest classification accuracy (98.90 %), Cohen-kappa score (0.941), F1-score (0.969), and the lowermost number of test loss as 0.102776 and MSE (0.0230) indicating that the model outperforms other HDL classifiers.
HIGHLIGHTS
- The potential of Raman spectroscopy in combination with hybrid deep learning (HDL) models to diagnose and classify cancerous or noncancerous samples, specifically blood plasma samples, based on chemical composition
- The implementation of data augmentation techniques to address underfitting and overfitting issues occur in the classification of spectral samples due to a lack of sufficient Raman spectral data
- The development of an efficient Hybrid Deep Learning (HDL) neural network model to differentiate breast cancer blood plasma from control samples and the use of spectral features as spectral cancer markers for breast cancer detection
- The evaluation of several HDL models for binary classification of Raman spectral signals, with the stacked 1D-CNN-GRU model achieving the highest classification accuracy and the lowest test losses
- The potential for this technique is to accurately classify breast cancerous samples and reduce the number of unnecessary excisional breast biopsies
GRAPHICAL ABSTRACT
Downloads
Metrics
References
F Bray, J Ferlay, I Soerjomataram, RL Siegel, LA Torre and A Jemal. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Canc. J. Clinicians 2018; 68, 394-424.
W Gordenne. Mammography: The gold standard of breast mass screening. J. Belg. Radiol. 1990; 73, 335-8.
HG Welch, LM Schwartz and S Woloshin. Ramifications of screening for breast cancer: 1 in 4 cancers detected by mammography are pseudocancers. BMJ 2006; 332, 727.
GW Auner, SK Koya, C Huang, B Broadbent, M Trexler, Z Auner, A Elias, KC Mehne and MA Brusatori. Applications of Raman spectroscopy in cancer diagnosis. Canc. Metastasis Rev. 2018; 37, 691-717.
R Manoharan, Y Wang and MS Feld. Histochemical analysis of biological tissues using Raman spectroscopy. Spectrochim. Acta Mol. Biomol. Spectros. 1996; 52, 215-49.
J Surmacki, J Musial, R Kordek and H Abramczyk. Raman imaging at biological interfaces: applications in breast cancer diagnosis. Mol. Canc. 2013; 12, 48.
AS Haka, KE Shafer-Peltier, M Fitzmaurice, J Crowe, RR Dasari and MS Feld. Identifying microcalcifications in benign and malignant breast lesions by probing differences in their chemical composition using Raman spectroscopy. Canc. Res. 2002; 62, 5375-80.
VS Renjith and PSH Jose. A noninvasive approach using multi-tier deep learning classifier for the detection and classification of breast neoplasm based on the staging of tumor growth. In: Proceedings of the 2020 International Conference on Decision Aid Sciences and Application, Sakheer, Bahrain. 2020, p. 471-8.
LA Austin, S Osseiran and CL Evans. Raman technologies in cancer diagnostics. Analyst 2016; 141, 476-503.
K Kong, C Kendall, N Stone and I Notingher. Raman spectroscopy for medical diagnostics-from in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 2015; 89, 121-34.
U Neugebauer, P Rösch and J Popp. Raman spectroscopy towards clinical application: Drug monitoring and pathogen identification. Int. J. Antimicrob. Agents 2015; 46, S35-9.
MM Joseph, N Narayanan, JB Nair, V Karunakaran, AN Ramya, PT Sujai, G Saranya, JS Arya, VM Vijayan and KK Maiti. Exploring the margins of SERS in practical domain: An emerging diagnostic modality for modern biomedical applications. Biomaterials 2018; 181, 140-81.
MJ Winterhalder and A Zumbusch. Beyond the borders-biomedical applications of non-linear raman microscopy. Adv. Drug Deliv. Rev. 2015; 89, 135-44.
G Agarwal, P Pradeep, V Aggarwal, CH Yip and PS Cheung. Spectrum of breast cancer in Asian women. World J. Surg. 2007; 31, 1031-40.
JRF Caldeira, ÉC Prando, FC Quevedo, FAM Neto, CA Rainho and SR Rogatto. CDH1 promoter hypermethylation and e-cadherin protein expression in infiltrating breast cancer. BMC Canc. 2006; 6, 48.
P Pradhan, S Guo, O Ryabchykov, J Popp and TW Bocklitz. Deep learning a boon for biophotonics? J. Biophotonics 2020; 13, e201960186.
MA Mohammed, B Al-Khateeb, AN Rashid, DA Ibrahim, MKA Ghani and SA Mostafa. Neural network and multi-fractal dimension features for breast cancer classification from ultrasound images. Comput. Electr. Eng. 2018; 70, 871-82.
RDH Devi and P Deepika. Performance comparison of various clustering techniques for diagnosis of breast cancer. In: Proceedings of the 2015 IEEE International Conference on Computational Intelligence and Computing Research, Madurai, India. 2015.
PJ Sudharshan, C Petitjean, F Spanhol, LE Oliveira, L Heutte and P Honeine. Multiple instances learning for histopathological breast cancer image classification. Expert Syst. Appl. 2019; 117, 103-11.
ZC Lipton, J Berkowitz and C Elkan. A critical review of recurrent neural networks for sequence learning. arXiv 2015; 1, 1-38.
J Dong, M Hong, Y Xu and X Zheng. A practical convolutional neural network model for discriminating Raman spectra of human and animal blood. J. Chemometr. 2019; 33, e3184
W Lee, ATM Lenferink, C Otto and HL Offerhaus. Classifying Raman spectra of extracellular vesicles based on convolutional neural networks for prostate cancer detection. J. Raman Spectros. 2020; 51, 293-300.
MK Maruthamuthu, AH Raffiee, DMD Oliveira, AM Ardekani, and MS Verma. Raman spectra-based deep learning: A tool to identify microbial contamination. MicrobiologyOpen 2020; 9, e1122.
S Boonsit, P Kalasuwan, PV Dommelen and C Daengngam. Rapid material identification via low-resolution Raman spectroscopy and deep convolutional neural network. J. Phys. Conf. 2021; 1719, 012081.
N Cheng, J Fu, D Chen, S Chen and H Wang. An antibody-free liver cancer screening approach based on nanoplasmonics biosensing chips via spectrum-based deep learning. NanoImpact 2021; 21, 100296.
J Houston, FG Glavin and MG Madden. Robust classification of high-dimensional spectroscopy data using deep learning and data synthesis. J. Chem. Inform. Model. 2020; 60, 1936-54.
CS Ho, N Jean, CA Hogan, L Blackmon, SS Jeffrey, M Holodniy, N Banaei, AAE Saleh, S Ermon and J Dionne. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Comm. 2019; 10, 4927.
J Ding, M Yu, L Zhu, T Zhang, J Xia and G Sun. Diverse spectral band-based deep residual network for tongue squamous cell carcinoma classification using fiber optic Raman spectroscopy. Photodiagnosis Photodynamic Ther. 2020; 32, 102048.
H Chen, C Chen, H Wang, C Chen, Z Guo, D Tong, H Li, H Li, R Si, H Lai and X Lv. Serum Raman spectroscopy combined with a multi-feature fusion convolutional neural network diagnosing thyroid dysfunction. Optik 2020; 216, 164961.
CM Krishna, GD Sockalingum, J Kurien, L Rao, L Venteo, M Pluot, M Manfait and VB Kartha. Micro-Raman spectroscopy for optical pathology of oral squamous cell carcinoma. Appl. Spectros. 2004; 58, 1128-35.
MS Bergholt, W Zheng and Z Huang. Characterizing variability in in vivo Raman spectroscopic properties of different anatomical sites of normal tissue in the oral cavity. J. Raman Spectros. 2012; 43, 255-62.
SP Singh, A Deshmukh and P Chaturvedi. In vivo Raman spectroscopic identification of premalignant lesions in oral buccal mucosa. J. Biomed. Optic. 2012; 17, 105002.
LP Choo-Smith, HGM Edwards, HP Endtz, JM Kros, F Heule, H Barr, JS Robinson, HA Bruining and GJ Puppels. Medical applications of Raman spectroscopy: From proof of principle to clinical implementation. Biopolymers 2002; 67, 1-9.
C Kallaway, LM Almond, H Barr, J Wood, J Hutchings, C Kendall and N Stone. Advances in the clinical application of Raman spectroscopy for cancer diagnostics. Photodiagnosis Photodynamic Ther. 2013; 10, 207-19.
P Zuvela, K Lin, C Shu, W Zheng, C Lim and Z Huang. Fiber-optic Raman spectroscopy with nature-inspired genetic algorithms enhances real-time in vivo detection and diagnosis of nasopharyngeal carcinoma. Anal. Chem. 2019; 91, 8101-8.
M Yu, H Yan, J Xia, L Zhu, T Zhang, Z Zhu, X Lou, G Sun and M Dong. Deep convolutional neural networks for tongue squamous cell carcinoma classification using Raman spectroscopy. Photodiagnosis and Photodynamic Ther. 2019; 26, 430-5.
J Xia, L Zhu, M Yu, T Zhang, Z Zhu, X Lou, G Sun and M Dong. Analysis and classification of oral tongue squamous cell carcinoma based on Raman spectroscopy and convolutional neural networks. J. Mod. Optic. 2020; 67, 481-9.
C Shu, H Yan, W Zheng, K Lin, A James, S Selvarajan, C Lim and Z Huang. Deep learning-guided fiberoptic raman spectroscopy enables real-time in vivo diagnosis and assessment of nasopharyngeal carcinoma and post-treatment efficacy during endoscopy. Anal. Chem. 2021; 93, 10898-906.
T Sciortino, R Secoli, E D’Amico, S Moccia, MC Nibali, L Gay, M Rossi, N Pecco, A Castellano, ED Momi, B Fernandes, M Riva and L Bello. Raman spectroscopy and machine learning for IDH genotyping of unprocessed glioma biopsies. Cancers 2021; 13, 4196.
M Riva, T Sciortino, R Secoli, E D’Amico, S Moccia, B Fernandes, MC Nibali, L Gay, M Rossi, ED Momi and L Bello. Glioma biopsies classification using raman spectroscopy and machine learning models on fresh tissue samples. Cancers 2021; 13, 1073.
K Mehta, A Atak, A Sahu and S Srivastava. An early investigative serum Raman spectroscopy study of meningioma. Analyst 2018; 143, 1916-23.
K Liua, B Liua, Y Zhang, Q Wu, M Zhong, L Shang, Y Wang, P Liang, W Wang, Q Zhao and B Li. Building an ensemble learning model for gastric cancer cell line classification via rapid raman spectroscopy. Comput. Struct. Biotechnol. J. 2023; 21, 802-11.
MS Silva-López, CAI Hernández, HRN Contreras, ÁGR Vázquez, A Ortiz-Dosal and ES Kolosovas-Machuca. Raman spectroscopy of individual cervical exfoliated cells in premalignant and malignant lesions. Appl. Sci. 2022; 12, 2419.
Z Liu, T Li, Z Wang, J Liu, S Huang, BH Min, JY An, KM Kim, S Kim, Y Chen, H Liu, Y Kim, DTW Wong, TJ Huang and X Ya-Hong. Gold nanopyramid arrays for non-invasive surface-enhanced raman spectroscopy-based gastric cancer detection via sEVs. ACS Appl. Nano Mater. 2022; 5, 12506-17.
L Huang, H Sun, L Sun, K Shi, Y Chen, X Ren, Y Ge, D Jiang, X Liu, W Knoll, Q Zhang and Y Wang. Rapid, label-free histopathological diagnosis of liver cancer based on Raman spectroscopy and deep learning. Nat. Comm. 2023; 14, 48.
M Jeng, LCT Sharma, S Huang, L Chang, S Wu and L Chow. Raman spectroscopy analysis for optical diagnosis of oral cancer detection. J. Clin. Med. 2019; 8, 1313.
C He, X Wu, J Zhou, Y Chen and J Ye. Raman optical identification of renal cell carcinoma via machine learning. Spectrochim. Acta Mol. Biomol. Spectros. 2021; 252, 119520.
C Chen, W Wu, C Chen, F Chen, X Dong, M Ma, Z Yan, X Lv, Y Ma and M Zhu. Rapid diagnosis of lung cancer and glioma based on serum Raman spectroscopy combined with deep learning. J. Raman Spectros. 2021; 52, 1798-809.
D Bury, G Faust, M Paraskevaidi, KM Ashton, TP Dawson and FL Martin. Phenotyping metastatic brain tumors applying spectrochemical analyses: Segregation of different cancer types. Anal. Lett. 2019; 52, 575-87.
K Aubertin, VQ Trinh, M Jermyn, P Baksic, AA Grosset, J Desroches, K St-Arnaud, M Birlea, MC Vladoiu, M Latour, R Albadine, F Saad, F Leblond and D Trudel. Mesoscopic characterization of prostate cancer using Raman spectroscopy: Potential for diagnostics and therapeutics. BJU Int. 2018; 122, 326-36.
H Yan, M Yu, J Xia, L Zhu, T Zhang, Z Zhu and G Sun. Diverse region-based CNN for tongue squamous cell carcinoma classification with Raman spectroscopy. IEEE Access 2020; 8, 127313-28.
X Wu, S Li, Q Xu, X Yan, Q Fu, X Fu, X Fang and Y Zhang. Rapid and accurate identification of colon cancer by Raman spectroscopy coupled with convolutional neural networks. Jpn. J. Appl. Phys. 2021; 60, 067001.
IP Santos, PV Doorn, PJ Caspers, TCB Schut, EM Barroso, TE Nijsten, VN Hegt, S Koljenovi´c and GJ Puppels. Improving clinical diagnosis of early-stage cutaneous melanoma based on Raman spectroscopy. Br. J. Canc. 2018; 119, 1339.
JD Gelder, KD Gussem, P Vandenabeele and L Moens. Reference database of Raman spectra of biological molecules. J. Raman Spectros. 2007; 38, 1133-47.
AD Meade, FM Lyng, P Knief and HJ Byrne. Growth substrate induced functional changes elucidated by FTIR and Raman spectroscopy in in-vitro cultured human keratinocytes. Anal. Bioanalytical Chem. 2007; 387, 1717-28.
I Notingher. Raman spectroscopy cell-based biosensors. Sensors 2007; 7, 1343-58.
P Jess, V Garcés-Chávez, D Smith, M Mazilu, L Paterson, A Riches, C Herrington, W Sibbett and K Dholakia. Dual beam fibre trap for Raman microspectroscopy of single cells. Optic. Express 2006; 14, 5779-91.
Z Movasaghi, S Rehman and IU Rehman. Raman spectroscopy of biological tissues. Appl. Spectros. Rev. 2007; 42, 493-541.
H Nawaz, F Bonnier, P Knief, O Howe, FM Lyng, AD Meade and HJ Byrne. Evaluation of the potential of Raman microspectroscopy for prediction of chemotherapeutic response to cisplatin in lung adenocarcinoma. Analyst 2010; 135, 3070-6.
MH Mozaffari and T Li-Lin. Overfitting one-dimensional convolutional neural networks for Raman spectra identification. Spectrochim. Acta Mol. Biomol. Spectros. 2022; 272, 120961.
A Shrivastava, T Pfister, O Tuzel, J Susskind, W Wang and R Webb. Learning from simulated and unsupervised images through adversarial training. In: Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, Hawaii. 2017.
I Goodfellow, Y Bengio and A Courville. Deep learning. MIT press: Cambridge, Massachusetts, 2016.
J Chen, X Hu and Y Liu. Improved gated recurrent units for time series prediction. In: Proceedings of the 2018 IEEE International Conference on Big Data, Seattle, Washington. 2018
Y Liu, S Chen and J Li. Deep learning for time series modelling. arXiv, Illinois, 2015.
MF Karim, MJ Hossain and A Svyatkovskiy. Multivariate time series forecasting using 1D convolutional neural networks. In: Proceedings of the 2019 International Conference on Computer and Communication Engineering, Pune, India. 2019.
DM Durairaj and BHK Mohan. A convolutional neural network-based approach to financial time series prediction. Neural Comput. Appl. 2022; 34, 13319-37.
X Wang, J Xu, W Shi and J Liu. OGRU: An optimized gated recurrent unit neural network. J. Phys. Conf. 2019; 1325, 012089.
J Du, Y Cheng, Q Zhou, J Zhang, X Zhang and G Li. Power load forecasting using BiLSTM-attention. IOP Conf. Ser. Earth Environ. Sci. 2020; 440, 032115.
D Gjylapi and E Proko. Recurrent neural networks in time series prediction. In: Proceedings of the New Research and Advances on Computer Science and Information Technology NRACSIT-2017, Vlora, Albania. 2017.
F Chollet. Xception: Deep Learning with Depthwise Separable Convolutions. In: Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, HI, USA. 2017, p. 1800-7.
M Abadi, P Barham, J Chen, Z Chen, A Davis, J Dean, M Devin, S Ghemawat, G Irving, M Isard, M Kudlur, J Levenberg, R Monga, S Moore, DG Murray, B Steiner, P Tucker, V Vasudevan, P Warden, M Wicke, Y Yu and X Zheng. TensorFlow: A system for large-scale machine learning. arXiv 2016; 1605, 08695.
DO Oyewola, EG Dada, S Misra and R Damaševicius. A novel data augmentation convolutional neural network for detecting malaria parasite in blood smear images. Appl. Artif. Intell. 2022; 36, 2033473.
DO Oyewola, EG Dada and O Ezekiel. Predicting nigerian stock returns using technical analysis and machine learning. Eur. J. Electr. Eng. Comput. Sci. 2019; 3, 1-8.
D Yao, B Li, H Liu, J Yang and L Jia. Remaining useful life prediction of roller bearings based on improved 1D-CNN and simple recurrent unit. Measurement 2021; 175, 109166.
M Wei, H Gu, M Ye, Q Wang, X Xu and C Wu. Remaining useful life prediction of lithium-ion batteries based on monte carlo dropout and gated recurrent unit. Energ. Rep. 2021; 7, 2862-71.
J Zhang, F Wei, F Feng and C Wang. Spatial-spectral feature refinement for hyperspectral image classification based on attention-dense 3D-2D-CNN. Sensors 2020; 20, 5191.
Y Zhang, M Liu, L Kong, T Peng, D Xie, L Zhang, L Tian and X Zou. Temporal characteristics of stress signals using GRU algorithm for heavy metal detection in rice based on sentinel-2 images. Int. J. Environ. Res. Publ. Health 2022; 19, 2567.
QJ Lv, HY Chen, WB Zhong, YY Wang, JY Song, SD Guo, LX Qi and CY Chen. A multi-task group Bi-LSTM networks application on electrocardiogram classification. IEEE J. Translational Eng. Health Med. 2019; 8, 1900111.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






