Characterization and Genetic Relationships of Vibrio spp. Isolated from Seafood in Retail Markets, Yala, Thailand
DOI:
https://doi.org/10.48048/tis.2023.5962Keywords:
V. cholerae, V. parahaemolyticus, Virulence genes, Hemolysin, Biofilm production, Genetic relationshipsAbstract
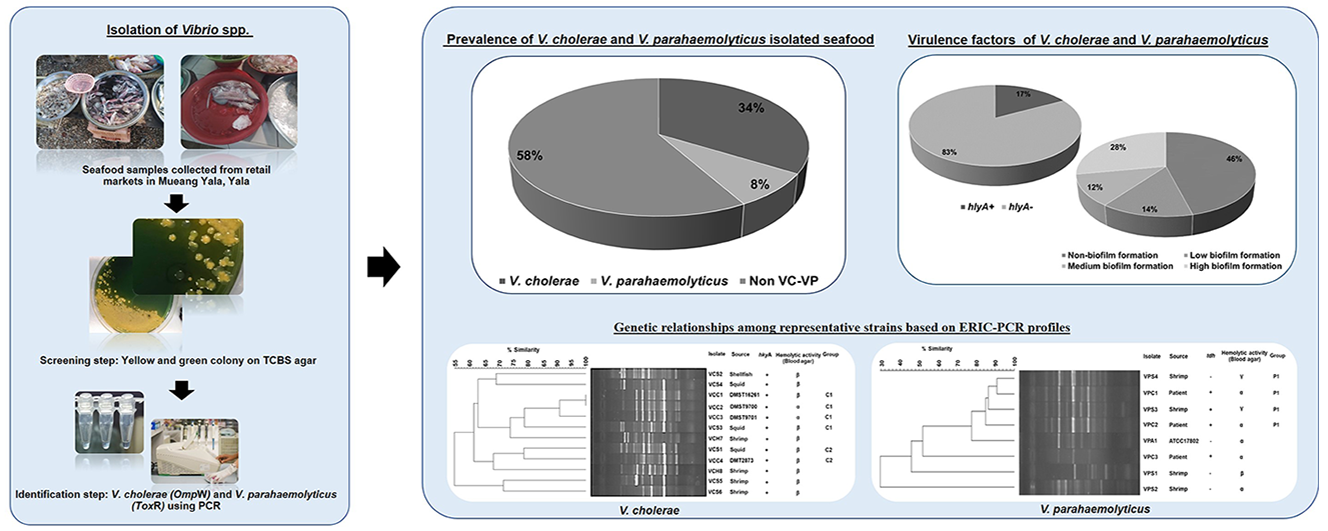
Vibrio spp. including Vibrio cholerae and V. parahaemolyticus are the major causative agent of diarrhea involved in seafood consumption. Yala is a southern border province in Thailand where no seafood sources are provided. Thus, it is necessary to import several seafood products from neighboring provinces. The risk of Vibrio spp. replication is increased during seafood transport. Therefore, the objectives of this study were to isolate and characterize V. cholerae and V. parahaemolyticus from seafood in Yala and to investigate their genetic relationships with virulent and clinical strains. A total of 100 Vibrio isolates were obtained from four different sample types in retail markets. It was found that two-thirds of all isolates were yellow colonies on TCBS agar. Using PCR with specific primers, 34 % of V. cholerae and 8 % of V. parahaemolyticus were identified. To classify the virulence strains, Vibrio spp. were positive for hlyA and tdh, 17 and 1 %, respectively. Moreover, 54 % of Vibrio isolates were able to produce biofilm and 28 % of those isolates showed a high level of biofilm production. All representative strains of V. cholerae presented ß-hemolysin on blood agar correlated to hlyA while representative strains of V. parahaemolyticus presented γ-hemolysin and ß-hemolysin on blood agar. In addition, ERIC-PCR provided different DNA profiles of representative V. cholerae and V. parahaemolyticus which some strains showed a correlation with clinical strains and virulent strains. Therefore, these findings might indicate the risk of Vibrio infections for seafood consumers, especially in Yala province where seafood needs to be imported from other provinces.
HIGHLIGHTS
- The first report showed high risk of Vibrio cholerae and V. parahaemolyticus spread in non-seafood source area: Yala province
- The majority of Vibrio cholerae and V. parahaemolyticus isolated from seafood in retail markets, Yala, Thailand was classified as virulent strains which possess major virulence genes, hemolytic activity, and biofilm formation
- Several Vibrio isolates obtained from seafood showed genetic similarities with virulent and clinical strains
GRAPHICAL ABSTRACT
Downloads
References
V Vuddhakul. Vibrio parahaemolyticus an important seafood-borne pathogen. iQue media, Songkla, Thailand, 2008, p. 221.
CL Tarr, JS Patel, ND Puhr, EG Sowers, CA Bopp and NA Strockbine. Identification of Vibrio isolates by a multiplex PCR assay and rpoB sequence determination. J. Clin. Microbiol. 2007; 45, 134-40.
FH Yildiz and KL Visick. Vibrio biofilms: So much the same yet so different. Trends Microbiol. 2009; 17, 109-18.
JB Kaper, JG Morris and MM Levine. Cholera. Clin. Microbiol. Rev. 1995; 8, 48-86.
P Figueroa-Arredondo, JE Heuser, NS Akopyants, JH Morisaki, S Giono-Cerezo, F Enrı́quez-Rincón and DE Berg. Cell vacuolation caused by Vibrio cholerae hemolysin. Infect. Immun. 2001; 69, 1613-24.
P Kissalai, S Preeprem, V Vuddhakul and P Mittraparp-arthorn. Phylogenetic analysis of atypical hemolysin gene in Vibrio campbellii and effects of cultivation salinity and pH on hemolytic activity and virulence. Walailak J. Sci. Tech. 2020; 17, 324-33.
BA Annous, PM Fratamico and JL Smith. Scientific status summary: Quorum sensing in biofilms: Why bacteria behave the way they do. J. Food Sci. 2009; 74, R24-R37.
KLV Dellen and PI Watnick. The Vibrio cholerae biofilm: A target for novel therapies to prevent and treat cholera. Drug Discov. Today Dis. Mech. 2006; 3, 261-6.
RS Mueller, D McDougald, D Cusumano, N Sodhi, S Kjelleberg, F Azam and DH Bartlett. Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J. Bacteriol. 2007; 189, 5348-60.
Annual Epidemiological Surveillance Report 2014, Available at: https://apps-doe.moph. go.th /boeeng/annual/Annual/AESR2014/index.php, accessed November 2021.
Annual Epidemiological Surveillance Report 2015, Available at: https://apps-doe.moph. go.th/boeeng/annual/Annual/AESR2015/, accessed November 2021.
Annual Epidemiological Surveillance Report 2017, Available at: https://apps-doe.moph. go.th/boeeng/download/AESR-6112-24.pdf, accessed November 2021.
Annual Epidemiological Surveillance Report 2018, Available at: https://apps.doe.moph. go.th/boeeng/download/AW_Annual_Mix%206212_14_r1.pdf, accessed November 2021.
Annual Epidemiological Surveillance Report 2019, Available at: https://apps-doe.moph. go.th/boeeng/download/MIX_AESR_2562.pdf, accessed November 2021.
Annual Epidemiological Surveillance Report 2020, Available at: https://apps-doe.moph. go.th/boeeng/download/AW_AESR_2563_MIX.pdf, accessed August 2022.
A Depaola, ML Motes and RM Mcphearson. Comparison of APHA and elevated temperature enrichment methods for recovery of Vibrio cholerae from oysters: Collaborative study. J. Assoc. Off. Anal. Chem. 1988; 71, 584-9.
CA Kaysner, A DePaola and J Jones. Vibrio, Available at: https://www.fda.gov/food/ laboratory-methods-food/bam-chapter-9-vibrio, accessed February 2021.
S Thaithongnum, P Ratanama, K Weeradechapol, A Sukhoom and V Vuddhakul. Detection of V. harveyi in shrimp postlarvae and hatchery tank water by the most probable number technique with PCR. Aquaculture 2006; 261, 1-9.
P Keim, A Klevytska, L Price, J Schupp, G Zinser, K Smith, M Hugh‐Jones, R Okinaka, K Hill, Jackson P. Molecular diversity in Bacillus anthracis. J Appl Microbiol. 1999; 87, 215-7.
B Nandi, RK Nandy, S Mukhopadhyay, GB Nair, T Shimada and AC Ghose. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 2000; 38, 4145-51.
YB Kim, J Okuda, C Matsumoto, N Takahashi, S Hashimoto and M Nishibuchi. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 1999; 37, 1173-7.
IN Rivera, J Chun, A Huq, RB Sack and RR Colwell. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microb. 2001; 67, 2421-9.
J Nesper, CM Lauriano, KE Klose, D Kapfhammer, A Kraiß and J Reidl. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: Influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 2001; 69, 435-45.
AK Singh, P Prakash, A Achra, GP Singh, A Das and RK Singh. Standardization and classification of in vitro biofilm formation by clinical isolates of Staphylococcus aureus. J. Glob. Infect. Dis. 2017; 9, 93.
R Buxton. Blood agar plates and hemolysis protocols. American Society for Microbiology. Washington DC, 2005, p. 1-9.
Y Zulkifli, M Alitheen, N Banu, S Radu, RA Rahim, S Lihan and S Yeap. Random amplified polymorphic DNA-PCR and ERIC PCR analysis on Vibrio parahaemolyticus isolated from cockles in Padang, Indonesia. Int. Food Res. J. 2009; 16, 141-50.
M Bonnin-Jusserand, S Copin, CL Bris, T Brauge, M Gay, A Brisabois, T Grard and G Midelet-Bourdin. Vibrio species involved in seafood-borne outbreaks (Vibrio cholerae, V. parahaemolyticus and V. vulnificus): Review of microbiological versus recent molecular detection methods in seafood products. Crc. Rev. Food Sci. 2019; 59, 597-610.
Y Nakaguchi. Contamination with Vibrio parahaemolyticus and its virulent strains in seafood marketed in Thailand, Vietnam, Malaysia, and Indonesia. Trop. Med. Health 2013; 41, 95-102.
X Song, J Zang, W Yu, X Shi and Y Wu. Occurrence and identification of pathogenic Vibrio contaminants in common seafood available in a chinese traditional market in qingdao, shandong province. Front. Microbiol. 2020; 11, 1488.
CW Tan, Rukayadi Y, Hasan H, Thung TY, Lee E, Rollon WD, Hara H, Kayali AY, Nishibuchi M, Radu S. Prevalence and antibiotic resistance patterns of Vibrio parahaemolyticus isolated from different types of seafood in Selangor, Malaysia. Saudi J Biol Sci. 2020; 27, 1602-8.
J Woodring, A Srijan, P Puripunyakom, W Oransathid, B Wongstitwilairoong and C Mason. Prevalence and antimicrobial susceptibilities of Vibrio, Salmonella, and Aeromonas isolates from various uncooked seafoods in Thailand. J. Food Prot. 2012; 75, 41-7.
M Ansaruzzaman, M Lucas, JL Deen, N Bhuiyan, XY Wang, A Safa, M Sultana, A Chowdhury, GB Nair and DA Sack. Pandemic Serovars (O3: K6 and O4: K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique: Pread of the pandemic into the African continent. 2005; 43, 2559-562.
YMG Hounmanou, P Leekitcharoenphon, RS Hendriksen, TV Dougnon, RH Mdegela, JE Olsen and A Dalsgaard. Surveillance and genomics of toxigenic Vibrio cholerae O1 from fish, phytoplankton and water in Lake Victoria, Tanzania. Front. Microbiol. 2019; 10, 901.
S Rao, WY Ngan, LC Chan, P Sekoai, AHY Fung, Y Pu, Y Yao and O Habimana. Questioning the source of identified non-foodborne pathogens from food-contact wooden surfaces used in Hong Kong's urban wet markets. One Health 2021; 13, 100300.
TTH Malcolm, WS Chang, YY Loo, YK Cheah, CWJWM Radzi, HK Kantilal, M Nishibuchi and R Son. Simulation of improper food hygiene practices: A quantitative assessment of Vibrio parahaemolyticus distribution. Int. J. Food Microbiol. 2018; 284, 112-19.
NT Popović, AB Skukan, Džidara P, Čož-Rakovac R, Strunjak-Perović I, Kozačinski L, Jadan M, Brlek-Gorski D. Microbiological quality of marketed fresh and frozen seafood caught off the Adriatic coast of Croatia. Veterinární Medicína 2010; 55, 233-41.
D Ottaviani, L Medici, G Talevi, M Napoleoni, P Serratore, E Zavatta, G Bignami, L Masini, S Chierichetti and S Fisichella. Molecular characterization and drug susceptibility of non-O1/O139 V. ácholerae strains of seafood, environmental and clinical origin, Italy. Food Microbiol. 2018; 72, 82-8.
S Preeprem, K Singkhamanan, M Nishibuchi, V Vuddhakul, P Mittraparp-arthorn. Multiplex multilocus variable-number tandem-repeat analysis for typing of pandemic Vibrio parahaemolyticus O1: KUT isolates. Foodborne Pathog. Dis. 2019; 16, 104-13.
S Preeprem, P Mittraparp-arthorn, P Bhoopong and V Vuddhakul. Isolation and characterization of Vibrio cholerae isolates from seafood in Hat Yai City, Songkhla, Thailand. Foodborne Pathog. Dis. 2014; 11, 881-6.
Y Hara-Kudo, K Sugiyama, M Nishibuchi, A Chowdhury, J Yatsuyanagi, Y Ohtomo, A Saito, H Nagano, T Nishina and H Nakagawa. Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3: K6 in seafood and the coastal environment in Japan. Appl. Environ. Microb. 2003; 69, 3883-91.
BM Rao and P Surendran. Genetic heterogeneity of non‐O1 and non‐O139 Vibrio cholerae isolates from shrimp aquaculture system: A comparison of RS‐, REP‐and ERIC‐PCR fingerprinting approaches. Lett. Appl. Microbiol. 2010; 51, 65-74.
DE Waturangi, I Joanito, Y Yogi and S Thomas. Use of REP-and ERIC-PCR to reveal genetic heterogeneity of Vibrio cholerae from edible ice in Jakarta, Indonesia. Gut. Pathog. 2012; 4, 1-9.
HA Ahmed, RS Abdelazim, RM Gharieb, RMA Elez and MA Awadallah. Prevalence of antibiotic resistant V. parahaemolyticus and V. cholerae in fish and humans with special reference to virulotyping and genotyping of V. parahaemolyticus. Slov. Vet. Res. 2018; 55, 251-62.
Downloads
Published
Issue
Section
License
Copyright (c) 2022 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






