Plant-derived Antiviral Compounds as Potential COVID-19 Drug Candidates: In-silico Investigation in Search of SARS-CoV-2 Inhibitors
DOI:
https://doi.org/10.48048/tis.2023.5529Keywords:
COVID-19, SARS-CoV-2, Antiviral, Molecular docking, ADMETAbstract
Corona virus disease 2019 (COVID-19) is an infectious disease caused by SARS-CoV-2, a newly discovered pathogenic corona virus that causes respiratory illness in humans and has now become a major challenge for the entire world. Although several vaccines for COVID-19 have been approved to date, lead compounds are still in high demand for the development of more promising or effective drugs and vaccines. According to the literature, antiviral drugs are used to treat COVID-19; thus, the current study is an attempt to screen lead molecules from plant-derived antiviral compounds. In this study, 33 plant-derived compounds with antiviral properties against corona viruses were used as ligand molecules, with Favilavir, Remdesivir, Ivermectin, and Dexamethasone serving as reference drugs. Molecular docking was performed between the selected ligands and the four main drug targets of SARS-CoV-2, Nucleocapsid Protein, Spike Glycoprotein, Proteases 3CLpro, and Helicase, and ADMET properties of screened compounds were observed. Docking results revealed that Reserpine, Tetrandrine, and Xanthoangelol F had a high Moldock score with each target, just like the current COVID-19 medications, Ivermectin and Remdesivir. Docking and ADMET studies indicate that Reserpine, Tetrandrine, and Xanthoangelol F may be potent lead compounds for the treatment of COVID-19 and should be investigated further.
HIGHLIGHTS
- Corona virus disease 2019 (COVID-19) is an infectious disease caused by the SARS-CoV-2 virus that poses a threat to humans
- Plant-derived drugs have showed significant antiviral activity against SARS-CoV, MERS-CoV29, HCoV-229E, and HCoV-OC43
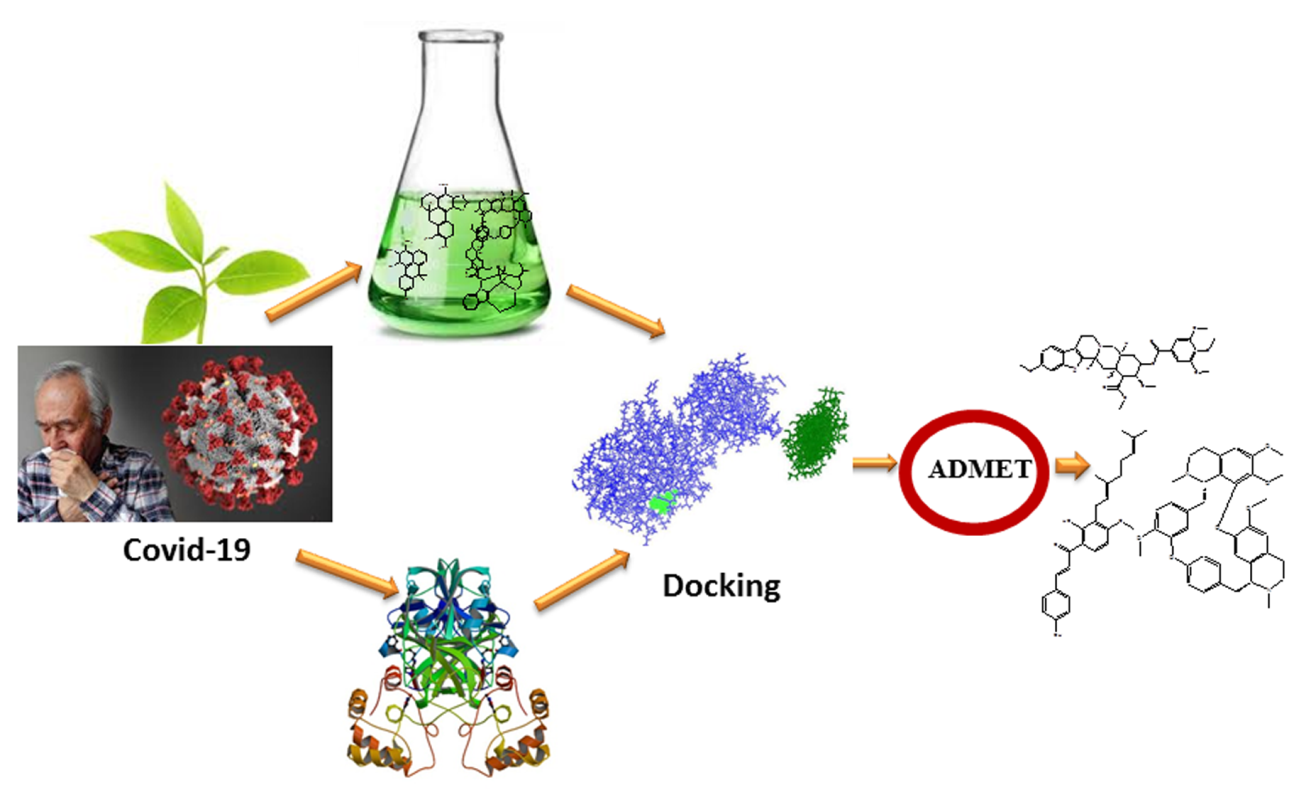
- The current study is an in-silico evaluation of plant-derived SARS-CoV-2 inhibitors. Molecular docking and hydrogen bond interaction studies were performed between the selected plant-derived compounds and vital targets of SARS-CoV-2 to evaluate antiviral efficacy against COVID-19, followed by an ADMET study of the screened ligands
- This in-silico study determined the antiviral potential of plant-derived compounds against SARS-CoV-2, of which Reserpine, Tetrandrine, and Xanthoangelol F may be potent lead compounds for the treatment of COVID-19
GRAPHICAL ABSTRACT
Downloads
Metrics
References
X Xu, P Chen, J Wang, J Feng, H Zhou, X Li, W Zhong and P Hao. Evolution of the novel Corona virus from the ongoing Wuhan outbreak and modeling of its spike protfein for risk of human transmission. Sci. Chin. Life Sci. 2020; 63, 457-60.
I Chakraborty and P Maity. COVID-19 outbreak: Migration, effects on society, global environment, and prevention. Sci. Total Environ. 2020; 728, 138882.
Weekly epidemiological update on COVID-19, Available at: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---31-august-2022, accessed August 2022.
J Cui, F Li and ZL Shi. Origin and evolution of pathogenic Corona viruses. Nat. Rev. Microbiol. 2019; 17, 181-92.
F Wu, S Zhao, B Yu,YM Chen, W Wang, ZG Song, Y Hu, ZW Tao, JH Tian, YY Pei, ML Yuan, YL Zhang, FH Dai, Y Liu, QM Wang, JJ Zheng, L Xu, E C Holmes and YZ Zhag. A new Corona virus associated with human respiratory disease in China. Nature 2020; 579, 265-9.
JF Chan, KH Kok, Z Zhu, H Chu, KK To, S Yuan and KY Yuen. Genomic characterization of the 2019 novel humanpathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020; 9, 221-36.
C Gil, T Ginex, I Maestro, V Nozal, L Barrado-Gil, MA Cuesta-Geijo, J Urquiza, D Ramírez, C Alonso, NE Campillo and A Martinez. COVID-19: Drug targets and potential treatments. J. Med. Chem. 2020; 63; 12359-86.
CK Chang, SC Lo, YS Wang and MH Hou. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today 2016; 21, 562-72.
S Kang, M Yang, Z Hong, L Zhang, Z Huang, X Chen, S He, Z Zhou, Z Zhou, Q Chen, Y Yan, C Zhang, H Shan and S Chen. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 2020; 10, 1228-38.
S Belouzard, JK Millet, BN Licitra and GR Whittaker. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012; 4, 1011-33.
D Wrapp, N Wang, KS Corbett, JA Goldsmith, CL Hsieh, O Abiona, BS Graham and JS McLellan. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020; 367, 1260-3.
R Hilgenfeld. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014; 281, 4085-96.
B Kolita, AJ Bora and P Hazarika. A Search of COVID-19 main protease inhibitor from plant derived alkaloids using chloroquine and hydroxychloroquine as reference: An in-silico approach. Int. J. Sci. Res. Biol. Sci. 2020; 7, 51-61.
Y Zhao, C Fang, Q Zhang, R Zhang, X Zhao, Y Duan, H Wang, Y Zhu, L Feng, J Zhao, M Shao, X Yang, L Zhang, CPeng, K Yang, D Ma, Z Rao and K Yang. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 2022; 13, 689-93.
KC Lehmann, EJ Snijder, CC Posthuma and AE Gorbalenya. What we know but do not understand about nidovirus helicases. Virus Res. 2015; 202, 12-32.
Z Jia, L Yan, Z Ren, L Wu, J Wang, JGuo, L Zheng, Z Ming, L Zhang, Z Lou and Z Rao. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon. Nucleic Acids Res. 2019; 47, 6538-50.
O Altay, E Mohammadi, S Lam, H Turkez, J Boren, J Nielsen, M Uhlen and A Mardinoglu. Current status of COVID-19 therapies and drug repositioning applications. iScience 2020; 23, 101303.
L Cynthia, Z Qiongqiong, L Yingzhu, LV Garner, SP Watkins, LJC Smoot, AC Gregg, AD Daniels, S Jervey and D Albaiu. Research and development on therapeutic agents and vaccines, for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020; 6, 315-31.
H Li, YM Wang, JY Xu and B Cao. Potential antiviral therapeutics for 2019 novel coronavirus. Chin. J. Tubercul. Respir. Dis. 2020; 43, E002.
ML Holshue, C DeBolt, S Lindquist, KH Lofy, J Wiesman, H Bruce, C Spitters, K Ericson, S Wilkerson, A Tural, G Diaz, A Cohn, LA Fox, A Patel, SI Gerber, L Kim, S Tong, X Lu, S Lindstrom, MA Pallansch, WC Weldon, …, SK Pillai. First case of 2019 novel coronavirus in the United States. New Engl. J. Med. 2020; 382, 929-36.
L Dong, S Hu and J Gao. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov. Ther. 2020; 14, 58-60.
PD Wakchaure, S Ghosh and BR Ganguly. Revealing the inhibition mechanism of RNA-dependent RNA polymerase (RdRp) of SARS-CoV‑2 by remdesivir and nucleotide analogues: A molecular dynamics simulation study. J. Phys. Chem. B 2020; 124, 10641-52.
L Caly, DJ Druce, MJ Catton, DA Jans and KM Wagstaff. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020; 178, 104787.
MA Pinzon, S Ortiz, H Holguın, JF Betancur, AD Cardona, H Laniado, CA Arias, B Muñoz, J Quiceno, D Jaramillo and Z Ramirez. Dexamethasone vs methylprednisolone high dose for Covid-19 pneumonia. PLoS One 2021; 16, e0252057.
OT Ebob, SB Babiaka and F Ntie-Kang. Natural products as potential lead compounds for drug discovery against SARS-CoV-2. Nat. Prod. Bioprospect. 2021; 11, 611-28.
CY Wu, JT Jan, SH Ma, CJ Kuo, HF Juan, YS Cheng, HH Hsu, HC Huang, D Wu, A Brik, FS Liang, RS Liu, JM Fang, ST Chen, PH Liang and CH Wong. Small molecules targeting severe acute respiratory syndrome human coronavirus. PNAS 2004; 10, 10012-7.
CC Wen, YH Kuo, JT Jan, PH Liang, SY Wang, HG Liu, CK Lee, ST Chang, CJ Kuo, SS Lee, CC Hou, PW Hsiao, SC Chien, LF Shyur and NS Yang. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007; 50, 4087-95.
CW Lin, FJ Tsai, CH Tsai, CC Lai, L Wan, TY Ho, CC Hsieh and PDL Chao. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005; 68, 36-42.
YB Ryu, HJ Jeong, JH Kim, YM Kim, JY Park, D Kim, TTH Nguyen, SJ Park, JS Chang, KH Park, MC Rho and WS Lee. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL (pro) inhibition. Bioorg. Med. Chem. 2010; 18, 7940-7.
PW Cheng, LT Ng, LC Chiang and CC Lin. Antiviral effects of saikosaponins on human coronavirus 229E in vitro.Clin. Exp. Pharmacol. Physiol. 2006; 33, 612-6.
DW Kim, KH Seo, MJ Curtis-Long, KY Oh, JW Oh, JK Cho, KH Lee and KH Park. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym Inhib. Med. Chem. 2014; 29, 59-63.
J Cinatl, B Morgenstern, G Bauer, P Chandra, H Rabenau and HW Doerr. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 2003; 361, 2045-6.
QY Yang, XY Tian and WS Fang. Bioactive coumarins from Boenninghauseniasessilicarpa. J. Asian Nat. Prod. Res. 2007; 9, 59-65.
SY Li, C Chen, HQ Zhang, HY Guo, H Wang, L Wang, X Zhang, SN Hua, J Yu, PG Xiao, RS Li and X Tan. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005; 67, 18-23.
JY Kim, YI Kim, SJ Park, IK Kim, YK Choi and SH Kim. Safe, high-throughput screening of natural compounds of MERS-CoV entry inhibitors using a pseudovirus expressing MERS-CoV spike protein. Int. J. Antimicrob. Agents 2018; 52, 730-2.
DE Kim, JS Min, MS Jang, JY Lee, YS Shin, JH Song, HR Kim, S Kim, YH Jin and S Kwon. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules 2019; 9, 696.
TY Ho, SL Wu, JC Chen, CC Li and CY Hsiang. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007; 74,92-101.
YB Ryu, SJ Park, YM Kim, JY Lee, WD Seo, JS Chang, KH Park, MC Rho and WS Lee. SARS-CoV 3CL pro inhibitory effects of quinone-methidetriterpenes from Tripterygiumregelii. Bioorg. Med. Chem. Lett. 2010; 20, 1873-6.
L Chen, J Li, C Luo, H Liu, W Xu, G Chen, OW Liew, W Zhu, CM Puah, X Shen and H Jiang. Binding interaction of quercetin-3-beta-galactoside and its synthetic derivatives with SARS-CoV3CL(pro): Structure-activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006; 14, 8295-306.
JY Park, JH Kim, YM Kim, HJ Jeong, DW Kim, KH Park, HJ Kwon, SJ Park, WS Lee and YB Ryu. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012; 20, 5928-35.
JY Park, HJ Jeong, JH Kim, YM Kim, SJ Park, D Kim, KH Park, WS Lee and YB Ryu. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012; 35, 2036-42.
MS Yu, J Lee, JM Lee, Y Kim, YW Chin, JG Jee, YS Keum, YJ Jeong. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett.2012; 22, 4049-54.
JY Park, JA Ko, DW Kim, YM Kim, HJ Kwon, HJ Jeong, CY Kim, KH Park, WS Lee and YB Ryu. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016; 31, 23-30.
R Thomsen and MH Christensen. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006; 49, 3315-21.
DK Gehlhaar, D Bouzida and PA Rejto. Fully automated and rapic flexible docking of inhibitors covalently bound to serine proteases. In: Proceedings of the 7th International Conference on Evolutionary Programming, California, United States. 1998.
JM Yang and CC Chen. GEMDOCK: A generic evolutionary method for molecular docking. Proteins 2004; 55, 288-304.
F Cheng, W Li, Y Zhou, J Shen, Z Wu, G Liu, PW Lee and Y Tang. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inform. Model. 2012; 52, 3099-105.
CA Lipinski, F Lombardo, BW Dominy and PJ Feeney. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997; 46, 3-26.
GA Jeffrey. An introduction to hydrogen bonding. Oxford University Press, Oxford, United Kingdom, 1992.
JA Williams, R Hyland, BC Jones, DA Smith, S Hurst, TC Goosen, V Peterkin, JR Koup and SE Ball. Drug-drug interactions for UDP-glucuronosyltransferase substrates: A pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab. Dispos. 2004; 32, 1201-8.
M Kacevska, GR Robertson, SJ Clarke and C Liddle. Inflammation and CYP3A4-mediated drug metabolism in advanced cancer: Impact and implications for chemotherapeutic drug dosing. Expet. Opin. Drug Metab. Toxicol. 2008; 4, 137-49.
KE Lasser, PD Allen, SJ Woolhandler and DU Himmelstein, SM Wolfe and DH Bor. Timing of new black box warnings and withdrawals for prescription medications. JAMA 2002; 287, 2215-20.
LC Wienkers, TG Heath. Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev. Drug Discov. 2005; 4, 825-33.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






