Molecular Modeling, Docking, and QSAR Studies on A Series of N-arylsulfonyl-N-2-pyridinyl-piperazines Analogs Acting as Anti-Diabetic Agents
DOI:
https://doi.org/10.48048/tis.2023.5472Keywords:
Glucokinase-Glucokinase regulatory protein (GK-GKRP/GCKR), Quantitative Structure Activity Relationship (QSAR), Docking, Multiple Linear Regression (MLR), Partial Least Square (PLS), Artificial Neural Network (ANN)Abstract
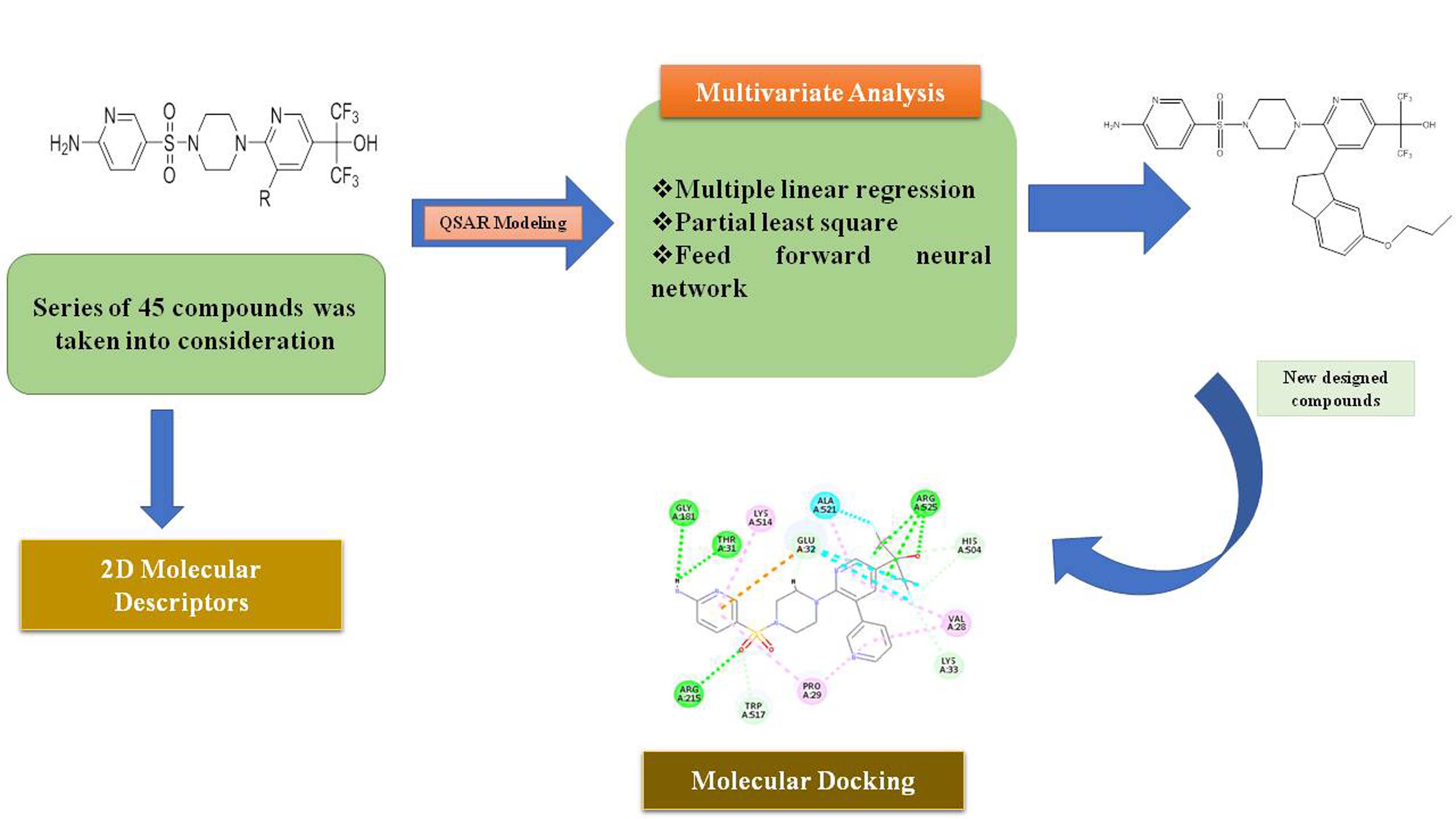
The Molecular structure of compounds contains a lot of information that can be used further. A classical Quantitative Structure Activity Relationship (QSAR) method was used to decode that information based on the descriptors. The study was performed on a mono-substituted series of Glucokinase-Glucokinase regulatory protein inhibitors (GK-GKRP/GCKR). A sequential application of the statistical method, both linear and nonlinear, has been used in the study which includes Multiple Linear Regression (MLR), Partial Least Square (PLS), and Artificial Neural Network (ANN). The developed model was validated using various statistical methods to evidently prove its reliability and precision. This knowledge will be used to design a new compound. Docking studies will be performed to establish the binding pattern of the designed compound. The prophetic power and robustness of the model containing 26 compounds in the training set were proven by the various statistical parameter s value: 0.30, F-value: 41.8, r: 0.94, r2: 0.88, r2CV: 0.77. The model gives insight into the various descriptors that are selected for the present study. The present study not only shows the contribution of various substituents in the biological activity but also indicates the changes that can be done to design the new potent molecules with more selectivity and less toxicity.
HIGHLIGHTS
- The enzyme glucokinase (GCK) is responsible for maintaining the body's normal glucose homeostasis. Hypertriglyceridemia, hyperinsulinemia, and T2D are all triggered by GCK dysfunction or dysregulation
- An inhibitor of the glucokinase enzyme (GCK), which is only present in hepatocytes and is in charge of glucose metabolism, is encoded by the glucokinase regulator (GCKR) gene
- The designed model gives insight into the various descriptors that are selected for the present study. The present study not only shows the contribution of various substituents in the biological activity but also indicates the changes that can be done to design the new potent molecules with more selectivity and less toxicity
- Small compounds have been discovered that specifically bind to GKRP and lower blood sugar levels
GRAPHICAL ABSTRACT
Downloads
References
FT Hong, MH Norman, KS Ashton, MD Bartberger, J Chen, S Chmait, R Cupples, C Fotsch, SR Jordan, DJ Lloyd and G Sivits. Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 4. Exploration of a novel binding pocket. J. Med. Chem. 2014; 57, 5949-64.
E Ermakova and R Kurbanov. Molecular insight into conformational transformation of human glucokinase: Conventional and targeted molecular dynamics. J. Biomol. Struct. Dynam. 2020; 38, 3035-45.
A Pautsch, N Stadler, A Löhle, W Rist, A Berg, L Glocker, H Nar, D Reinert, M Lenter, A Heckel and G Schnapp. Crystal structure of glucokinase regulatory protein. Biochemistry 2013; 52, 3523-31.
K Chen, K Michelsen, RJM Kurzeja, J Han, M Vazir, DJSJ Jr, C Hale and RC Wahl. Discovery of Small-Molecule Glucokinase Regulatory Protein Modulators That Restore Glucokinase Activity. SLAS Discov. 2014; 19, 1014-23.
H Watanabe, Y Inaba, K Kimura, M Matsumoto, S Kaneko, M Kasuga and H Inoue. Sirt2 facilitates hepatic glucose uptake by deacetylating glucokinase regulatory protein. Nat. Comm. 2018; 9, 30.
LD Mendelsohn. ChemDraw 8 Ultra, windows and macintosh versions. J. Chem. Inform. Model. 2004; 44, 2225-6.
Icon Group Ltd. Oxford Molecular Group PLC: International competitive benchmarks and financial gap analysis (financial performance series). Icon Group International, Inc. California, 2004.
A Dalby, JG Nourse, WD Hounshell, AK Gushurst, DL Grier, BA Leland and J Laufer. Description of several chemical structure file formats used by computer programs developed at molecular design limited. J. Chem. Inform. Comput. Sci. 1992; 32, 244-55.
A Nagpal and S Paliwal. Discovery of novel and selective c-jun nh2-terminal kinases 2 inhibitors by two-dimensional quantitative structure-activity relationship model development, molecular docking and absorption, distribution, metabolism, elimination prediction studies: An in silico approach. Asian J. Pharmaceut. Clin. Res. 2018; 11, 100-8.
R Arya, SP Gupta, S Paliwal, S Kesar, A Mishra and YS Prabhakar. QSAR and molecular modeling studies on a series of pyrrolidine analogs acting as BACE-1 Inhibitors. Lett. Drug Des. Discov. 2019; 16, 746-60.
K Roy and AS Mandal. Development of linear and nonlinear predictive QSAR models and their external validation using molecular similarity principle for anti-HIV indolyl aryl sulfones. J. Enzym. Inhib. Med. Chem. 2008; 23, 980-95.
L Saghaie, H Sakhi, H Sabzyan, M Shahlaei and D Shamshirian. Stepwise MLR and PCR QSAR study of the pharmaceutical activities of antimalarial 3-hydroxypyridinone agents using B3LYP/6-311++ G** descriptors. Med. Chem. Res. 2013; 22, 1679-88
H Kubinyi. QSAR and 3D QSAR in drug design part 1: Methodology. Drug Discov. Today 1997; 2, 457-67.
A Nagpal and SK Paliwal. QSAR model development using MLR, PLS and NN approach to elucidate the physicochemical properties responsible for neurologically important JNK3 inhibitory activity. J. Pharmaceut. Sci. Res. 2017; 9, 1831-43.
D Kim, SI Hong and DS Lee. The quantitative structure-mutagenicity relationship of polycylic aromatic hydrocarbon metabolites. Int. J. Mol. Sci. 2006; 7, 556-70.
A Tropsha and A Golbraikh. Predictive QSAR modeling workflow, model applicability domains, and virtual screening. Curr. Pharmaceut. Des. 2007; 13, 3494-504.
A Tropsha, P Gramatica and VK Gombar. The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Combin. Sci. 2003; 22, 69-77.
N Tripathi, S Paliwal, S Sharma, K Verma, R Gururani, A Tiwari, A Verma, M Chauhan, A Singh, D Kumar and A Pant. Discovery of novel soluble epoxide hydrolase inhibitors as potent vasodilators. Sci. Rep. 2018; 8, 14604.
LD Pennington, MD Bartberger, MD Croghan, KL Andrews, KS Ashton, MP Bourbeau, J Chen, S Chmait, R Cupples, C Fotsch, J Helmering, H Fang-Tsao, RW Hungate, SR Jordan, K Kong, L Liu, K Michelsen, C Moyer, N Nishimura, MH Norman, ..., DJSJ Jr. Discovery and structure-guided optimization of diarylmethanesulfonamide disrupters of glucokinase-glucokinase regulatory protein (GK-GKRP) binding: strategic use of a N→ S (nN→ σ* S–X) interaction for conformational constraint. J. Med. Chem. 2015; 58, 9663-79.
MO Taha, M Habash and MA Khanfar. The use of docking-based comparative intermolecular contacts analysis to identify optimal docking conditions within glucokinase and to discover of new GK activators. J. Comput. Aided Mol. Des. 2014; 28, 509-47.
T Beck and BG Miller. Structural basis for regulation of human glucokinase by glucokinase regulatory protein. Biochemistry 2013; 52, 6232-9.
EI Edache, A Uzairu, PA Mamza and AG Shallangwa. 2D-QSAR, 3D-QSAR, molecular docking, and molecular dynamics simulations in the probe of novel type I diabetes treatment. Int. J. New Chem. 2021, https://doi.org/10.22034/ijnc.2021.529335.1161
S Jain, A Bisht, K Verma, S Negi, S Paliwal and S Sharma. The role of fatty acid amide hydrolase enzyme inhibitors in Alzheimer’s disease. Cell Biochem. Funct. 2022; 40, 106-17.
Downloads
Published
Issue
Section
License
Copyright (c) 2022 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






