Prediction of Cytotoxicity Against HepG2 by Quantitative Structure-Activity Relation (QSAR) Modelling
DOI:
https://doi.org/10.48048/tis.2023.5388Keywords:
QSAR, Cancer, Cytotoxicity, Desirability, Virtual screening, Machine learning, ModelAbstract
Hepatocellular carcinoma (HCC) is the dominant subtype of liver cancer with very low survival rate but the chemotherapy for HCC is still in grey zone due to the limited efficacy and high toxicity profile of approved drugs raising the heavy demand on drug development for HCC. The study aimed to establish a desirability based quantitative structure activity relation (QSAR) model to predict the activity of chemical compounds against one of HCC cell line (HepG2). Different support vector machine (SVM) models were constructed and ensembled to 10 virtual screening protocols. These protocols were validated by an external dataset in combination with decoys as interference. Results showed that ensemble models exhibited improved area under the Receiver Operating Characteristic Curve (ROC), sensitivity, and specificity compared to base models in training and test set. When being validated for virtual screening to recover known active molecules in the mixture with known inactive and decoy compounds, all virtual screening protocols have good performance with good Boltzmann-Enhanced Discrimination of ROC (BEDROC) and enrichment factor (EF). The best protocol with BEDROC of 0.63 and EF of 29.55 was suitable for further screening of active compounds against HepG2 cell line.
HIGHLIGHTS
- Ensemble quantitative structure-activity relationship (QSAR) models improved the performance of single models
- Cytotoxicity prediction was transformed to desirability score as a general output for easily integration in multi-objective optimization
- Virtual models for screening of HepG2 cytotoxicity compounds were well validated
GRAPHICAL ABSTRACT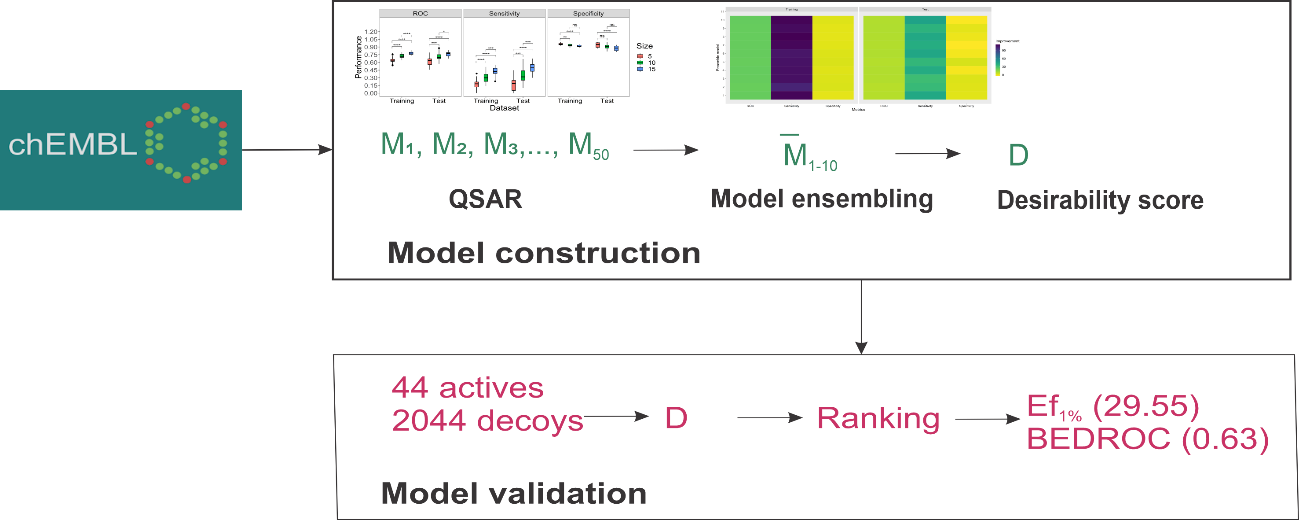
Downloads
Metrics
References
A Koulouris, C Tsagkaris, V Spyrou, E Pappa, A Troullinou and M Nikolaou. Hepatocellular carcinoma: An overview of the changing landscape of treatment options. J. Hepatocellular Carcinoma 2021; 8, 387-401.
EN Muratov, J Bajorath, RP Sheridan, IV Tetko, D Filimonov, V Poroikov, TI Oprea, II Baskin, A Varnek, A Roitberg, O Isayev, S Curtalolo, D Fourches, Y Cohen, A Aspuru-Guzik, DA Winkler, D Agrafiotis, A Cherkasov and A Tropsha. QSAR without borders. Chem. Soc. Rev. 2020; 49, 3525-64.
M Cruz-Monteagudo, F Borges and MNDS Cordeiro. Desirability-based multiobjective optimization for global QSAR studies: Application to the design of novel NSAIDs with improved analgesic, antiinflammatory, and ulcerogenic profiles. J. Comput. Chem. 2008; 29, 2445-59.
CA Coello, SG Brambila, JF Gamboa, MGC Tapia and RH Gómez. Evolutionary multiobjective optimization: Open research areas and some challenges lying ahead. Complex Intell. Syst. 2020; 6, 21-36.
VV Kleandrova, MT Scotti, L Scotti, A Nayarisseri and A Speck-Planche. Cell-based multi-target QSAR model for design of virtual versatile inhibitors of liver cancer cell lines. SAR QSAR Environ. Res. 2020; 31, 815-36.
NM O’Boyle, M Banck, CA James, C Morley, T Vandermeersch and GR Hutchison. Open babel: An open chemical toolbox. J. Cheminformatics 2011; 3, 33.
D Castillo-González, JL Mergny, AD Rache, G Pérez-Machado, MA Cabrera-Pérez, O Nicolotti, A Introcaso, GF Mangiatordi, A Guédin, A Bourdoncle, T Garrigues, F Pallardó, MNDS Cordeiro, C Paz-y-Miño, E Tejera, F Borges and M Cruz-Monteagudo. Harmonization of QSAR best practices and molecular docking provides an efficient virtual screening tool for discovering new G-quadruplex ligands. J. Chem. Inform. Model. 2015; 55, 2094-110.
Y Perez-Castillo, A Sánchez-Rodríguez, E Tejera, M Cruz-Monteagudo, F Borges, MNDS Cordeiro, H Le-Thi-Thu and H Pham-The. A desirability-based multi objective approach for the virtual screening discovery of broad-spectrum anti-gastric cancer agents. PLos One 2018; 13, e0192176.
F Sahigara, K Mansouri, D Ballabio, A Mauri, V Consonni and R Todeschini. Comparison of different approaches to define the applicability domain of QSAR models. Molecules 2012; 17, 4791-810.
R Polikar. Ensemble based systems in decision making. IEEE Circ. Syst. Mag. 2006; 6, 21-45.
MM Mysinger, M Carchia, JJ Irwin and BK Shoichet. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012; 55, 6582-94.
JF Truchon and CI Bayly. Evaluating virtual screening methods: Good and bad metrics for the “Early recognition” problem. J. Chem. Inf. Model. 2007; 47, 488-508.
S Kwon, H Bae, J Jo and S Yoon. Comprehensive ensemble in QSAR prediction for drug discovery. BMC Bioinformatics 2019; 20, 521.
P Pradeep, RJ Povinelli, S White and SJ Merrill. An ensemble model of QSAR tools for regulatory risk assessment. J. Cheminform. 2016; 8, 48.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






