Molecular Docking and Molecular Dynamics Simulation-Based Identification of Natural Inhibitors against Druggable Human Papilloma Virus Type 16 Target
DOI:
https://doi.org/10.48048/tis.2023.4891Keywords:
E5 protein, HPV16, Molecular dynamics, Ligand conformation, Ligand movementAbstract
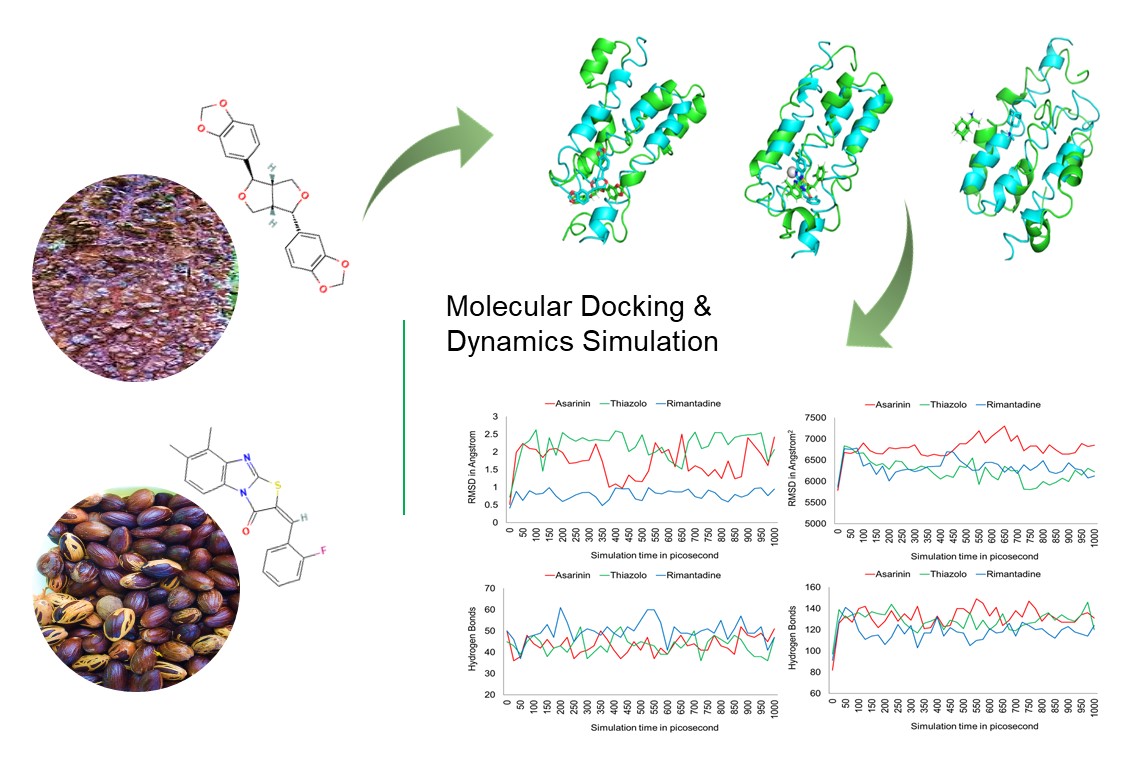
The E5 protein is the smallest known oncoprotein linked to HPV 16 cancer development. In this study, we determined the potential of asarinin and thiazolo as an inhibitor of the E5 protein through molecular dynamics. The results showed that the binding site is unstable because of its hydrophobic nature and small size, causing considerable changes in the binding site for each of the 3 drugs examined. Except for asarinin, which still interacts with the first hydrophobic domain, they preserved their capacity to prevent endosomal acidification, hyper amplification of the EGFR pathway and contact with BAP31. It may inhibit E5-MHC I interaction. Thiazolo[3,2-a]benzymidazole-3(2H)-one,2-(2-fluorobenzylideno)-7,8-dimethyl (thiazolo) is expected to form more stable protein-ligand complexes than the other 2. However, the SASA, hydrogen bond and DCCM plots show that both compounds are equivalent to HPV 16 E5 protein.
HIGHLIGHTS
- The smallest known oncoprotein associated with cancer development as a result of HPV 16 infection is the E5 protein
- The SASA, hydrogen bond, and DCCM plots, on the other hand, show that asarinin and thiazolo are equivalent to the HPV 16 E5 protein
- Thiazolo is predicted to produce more stable protein-ligand complexes than the others because of its unique molecular dynamic properties
GRAPHICAL ABSTRACT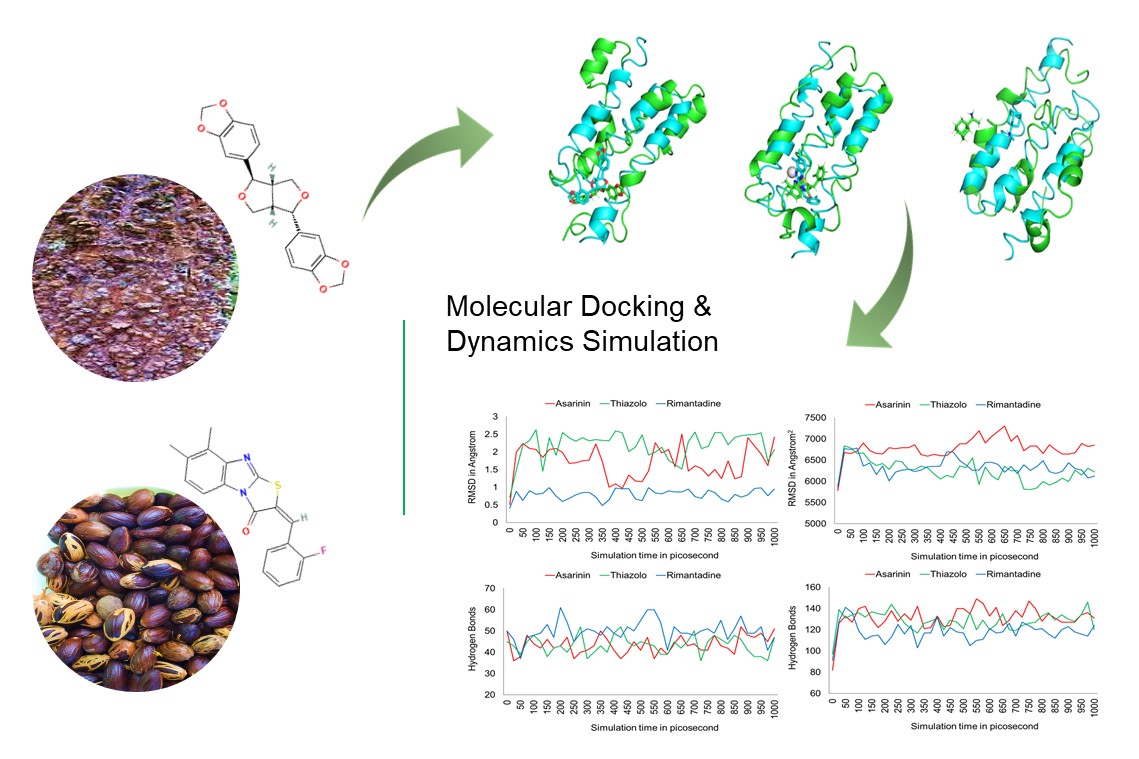
Downloads
References
M Tao, M Kruhlak, S Xia, E Androphy and ZM Zheng. Signals that dictate nuclear localization of human papillomavirus type 16 oncoprotein E6 in living cells. J. Virol. 2003; 77, 13232-47.
M Forcier and N Musacchio. An overview of human papillomavirus infection for the dermatologist: Disease, diagnosis, management, and prevention. Dermatol. Ther. 2010; 23, 458-76.
W Bonnez. Chapter 26 - human papillomavirus. In: ADT Barrett and LR Stanberry (Eds.). Vaccines for biodefense and emerging and neglected diseases. Academic Press, London, 2009, p. 469-96.
D Bzhalava, P Guan, S Franceschi, J Dillner and G Clifford. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 2013; 445, 224-31.
SV Graham. The human papillomavirus replication cycle, and its links to cancer progression: A comprehensive review. Clin. Sci. 2017; 131, 2201-21.
F Cobo. Introduction and epidemiological data. In: F Cobo (Ed.). Human papillomavirus infections. Woodhead Publishing, Cambridge, 2012, p. 1-14.
N Egawa and J Doorbar. The low-risk papillomaviruses. Virus Res. 2017; 231, 119-27.
K Kobayashi, K Hisamatsu, N Suzui, A Hara, H Tomita and T Miyazaki. A review of HPV-related head and neck cancer. J. Clin. Med. 2018; 7, 241.
S Miyauchi, PD Sanders, K Guram, SS Kim, F Paolini, A Venuti, EEW Cohen, JS Gutkind, JA Califano and AB Sharabi. HPV16 E5 mediates resistance to PD-L1 blockade and can be targeted with rimantadine in head and neck cancer. Canc. Res. 2020; 80, 732-46.
LF Wetherill, KK Holmes, M Verow, M Müller, G Howell, M Harris, C Fishwick, N Stonehouse, R Foster, GE Blair, S Griffin and A Macdonald. High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors. J. Virol. 2012; 86, 5341-51.
ADR Nurcahyanti. Cervical cancer: The case in Indonesia and natural product based therapy. J. Canc. Biol. Res. 2016; 4, 1078.
EJ Domingo, R Noviani, MR Noor, CA Ngelangel, KK Limpaphayom, TV Thuan, KS Louie and MA Quinng. Epidemiology and prevention of cervical cancer in Indonesia, Malaysia, the Philippines, Thailand and Vietnam. Vaccine 2008; 26, M71-9.
PV Chin-Hong and GE Reid. Human papillomavirus infection in solid organ transplant recipients: Guidelines from the American society of transplantation infectious diseases community of practice. Clin. Transplant. 2019; 33, e13590.
C Chelimo, TA Wouldes, LD Cameron and JM Elwood. Risk factors for and prevention of human papillomaviruses (HPV), genital warts and cervical cancer. J. Infect. 2013; 66, 207-17.
M Yamaguchi. Risk factors for HPV infection and high-grade cervical disease in sexually active Japanese women. Sci. Rep. 2021; 11, 2898.
WK Huh, EA Joura, AR Giuliano, OE Iversen, RP Andrade, KA Ault, D Bartholomew, RM Cestero, EN Fedrizzi, AL Hirschberg, MH Mayrand, AM Ruiz-Sternberg, JT Stapleton, DJ Wiley, A Ferenczy, R Kurman, BM Ronnett, MH Stoler, J Cuzick, …, A Luxembourg. Final efficacy, immunogenicity, and safety analyses of a nine-valent human papillomavirus vaccine in women aged 16-26 years: A randomised, double-blind trial. Lancet 2017; 390, 2143-59.
M El-Zein, L Richardson and EL Franco. Cervical cancer screening of HPV vaccinated populations: Cytology, molecular testing, both or none. J. Clin. Virol. 2016; 76, S62-8.
KE Gallagher, DS LaMontagne and D Watson-Jones. Status of HPV vaccine introduction and barriers to country uptake. Vaccine 2018; 36, 4761-7.
DM Harper and LR DeMars. HPV vaccines - a review of the first decade. Gynecol. Oncol. 2017; 146, 196-204.
E Kumakech, S Andersson, H Wabinga, C Musubika, S Kirimunda and V Berggren. Cervical cancer risk perceptions, sexual risk behaviors and sexually transmitted infections among bivalent human papillomavirus vaccinated and non-vaccinated young women in Uganda - 5 year follow up study. BMC Wom. Health 2017; 17, 40.
M Toots, M Ustav, A Männik, K Mumm, K Tämm, T Tamm, E Ustav and M Ustav. Identification of several high-risk HPV inhibitors and drug targets with a novel high-throughput screening assay. Plos Pathogens 2017; 13, e1006168.
J Durzynska, K Lesniewicz and E Poreba. Human papillomaviruses in epigenetic regulations. Mutat. Res. Rev. Mutat. Res. 2017; 772, 36-50.
DA Haręża, JR Wilczyński and E Paradowska. Human papillomaviruses as infectious agents in gynecological cancers. Oncogenic properties of viral proteins. Int. J. Mol. Sci. 2022; 23, 1818.
A Pal and R Kundu. Human papillomavirus E6 and E7: The cervical cancer hallmarks and targets for therapy. Front. Microbiol. 2020; 10, 3116.
NSL Yeo-Teh, Y Ito and S Jha. High-risk human papillomaviral oncogenes E6 and E7 target key cellular pathways to achieve oncogenesis. Int. J. Mol. Sci. 2018; 19, 1706.
J Doorbar, W Quint, L Banks, IG Bravo, M Stoler, TR Broker and MA Stanley. The biology and life-cycle of human papillomaviruses. Vaccine 2012; 30, F55-70.
A Venuti, F Paolini, L Nasir, A Corteggio, S Roperto, MS Campo and G Borzacchiello. Papillomavirus E5: The smallest oncoprotein with many functions. Mol. Canc. 2011; 10, 140.
D DiMaio and L Petti. The E5 proteins. Virology 2013; 445, 99-114.
M Gruener, IG Bravo, F Momburg, A Alonso and P Tomakidi. The E5 protein of the human papillomavirus type 16 down-regulates HLA-I surface expression in calnexin-expressing but not in calnexin-deficient cells. Virol. J. 2007; 4, 116.
R Nath, CA Mant, B Kell, J Cason and JM Bible. Analyses of variant human papillomavirus type-16 E5 proteins for their ability to induce mitogenesis of murine fibroblasts. Canc. Cell Int. 2006; 6, 19.
MI Rodríguez, ME Finbow and A Alonso. Binding of human papillomavirus 16 E5 to the 16 kDa subunit c (proteolipid) of the vacuolar H + −ATPase can be dissociated from the E5-mediated epidermal growth factor receptor overactivation. Oncogene 2000; 19, 3727-32.
GH Ashrafi, M Haghshenas, B Marchetti and MS Campo. E5 protein of human papillomavirus 16 downregulates HLA class I and interacts with the heavy chain via its first hydrophobic domain. Int. J. Canc. 2006; 119, 2105-12.
JA Regan and LA Laimins. Bap31 is a novel target of the human papillomavirus E5 protein. J. Virol. 2008; 82, 10042-51.
D Neofytos, C El-Beyrouty and JA DeSimone. Chapter 12 - pharmacobiology of infections. In: SA Waldman and A Terzic (Eds.). Pharmacology and therapeutics: Principles to practice. W. B. Saunders, Philadelphia, 2009, p. 173-91.
EA Govorkova, HB Fang, M Tan and RG Webster. Neuraminidase inhibitor-rimantadine combinations exert additive and synergistic anti-influenza virus effects in MDCK cells. Antimicrob. Agents Chemother. 2004; 48, 4855-63.
TL Foster, GS Thompson, AP Kalverda, J Kankanala, M Bentham, LF Wetherill, J Thompson, AM Barker, D Clarke, M Noerenberg, AR Pearson, DJ Rowlands, SW Homans, M Harris, R Foster and S Griffin . Structure-guided design affirms inhibitors of hepatitis C virus p7 as a viable class of antivirals targeting virion release. Hepatology 2014; 59, 408-22.
LF Wetherill, CW Wasson, G Swinscoe, D Kealy, R Foster, S Griffin and A Macdonald. Alkyl-imino sugars inhibit the pro-oncogenic ion channel function of human papillomavirus (HPV) E5. Antivir. Res. 2018; 158, 113-21.
MS Campo, SV Graham, MS Cortese, GH Ashrafi, EH Araibi, ES Dornan, K Miners, C Nunes and S Man. HPV-16 E5 down-regulates expression of surface HLA class I and reduces recognition by CD8 T cells. Virology 2010; 407, 137-42.
F Abe, NV Prooyen, JJ Ladasky and M Edidin. Interaction of Bap31 and MHC class I molecules and their traffic out of the endoplasmic reticulum. J. Immunol. 2009; 182, 4776-83.
M Müller, EL Prescott, CW Wasson and A Macdonald. Human papillomavirus E5 oncoprotein: Function and potential target for antiviral therapeutics. Future Virol. 2015; 10, 27-39.
KK Halavaty, J Regan, K Mehta and L Laimins. Human papillomavirus E5 oncoproteins bind the A4 endoplasmic reticulum protein to regulatze proliferative ability upon differentiation. Virology 2014; 452-453, 223-30.
HTT Lai, A Giorgetti, G Rossetti, TT Nguyen, P Carloni and A Kranjc. The interplay of cholesterol and ligand binding in hTSPO from classical molecular dynamics simulations. Molecules 2021; 26, 1250.
A Gutteridge and J Thornton. Conformational change in substrate binding, catalysis and product release: An open and shut case? FEBS Lett. 2004; 567, 67-73.
EC Hulme and MA Trevethick. Ligand binding assays at equilibrium: Validation and interpretation. Br. J. Pharmacol. 2010; 161, 1219-37.
I Aier, PK Varadwaj and U Raj. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 2016; 6, 34984.
KS Chopdar, GC Dash, PK Mohapatra, B Nayak and MK Raval. Monte-carlo method-based QSAR model to discover phytochemical urease inhibitors using SMILES and GRAPH descriptors. J. Biomol. Struct. Dyn. 2022; 40, 5090-9.
J Zhu, Y Lv, X Han, D Xu and W Han. Understanding the differences of the ligand binding/unbinding pathways between phosphorylated and non-phosphorylated ARH1 using molecular dynamics simulations. Sci. Rep. 2017; 7, 12439.
C Scott and S Griffin. Viroporins: Structure, function and potential as antiviral targets. J. Gen. Virol. 2015; 96, 2000-27.
J Kalikka and J Akola. Steered molecular dynamics simulations of ligand-receptor interaction in lipocalins. Eur. Biophys. J. 2011; 40, 181-94.
C Schmidt, JA Macpherson, AM Lau, KW Tan, F Fraternali and A Politis. Surface accessibility and dynamics of macromolecular assemblies probed by covalent labeling mass spectrometry and integrative modeling. Anal. Chem. 2017; 89, 1459-68.
JA Marsh and SA Teichmann. Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure 2011; 19, 859-67.
M Candotti, A Pérez, C Ferrer-Costa, M Rueda, T Meyer, JL Gelpí and M Orozco. Exploring early stages of the chemical unfolding of proteins at the proteome scale. Plos Comput. Biol. 2013; 9, e1003393.
M Tarek and DJ Tobias. The role of protein-solvent hydrogen bond dynamics in the structural relaxation of a protein in glycerol versus water. Eur. Biophys. J. 2008; 37, 701-9.
D Zhang and R Lazim. Application of conventional molecular dynamics simulation in evaluating the stability of apomyoglobin in urea solution. Sci. Rep. 2017; 7, 44651.
P Nagy. Competing intramolecular vs. intermolecular hydrogen bonds in solution. Int. J. Mol. Sci. 2014; 15, 19562-633.
M Petukhov, G Rychkov, L Firsov and L Serrano. H-bonding in protein hydration revisited. Protein Sci. 2004; 13, 2120-9.
SNH Ishak, SNAM Aris, KBA Halim, MSM Ali, TC Leow, NHA Kamarudin, M Masomian and RNZRA Rahman. Molecular dynamic simulation of space and earth-grown crystal structures of thermostable T1 lipase geobacillus zalihae revealed a better structure. Molecules 2017; 22, 1574.
I Chikalov, P Yao, M Moshkov and JC Latombe. Learning probabilistic models of hydrogen bond stability from molecular dynamics simulation trajectories. BMC Bioinformatics 2011; 12, S34.
JL England and G Haran. Role of solvation effects in protein denaturation: From thermodynamics to single molecules and back. Ann. Rev. Phys. Chem. 2011; 62, 257-77.
RPP Valente, RC Souza, MG Medeiros, JEV Ferreira, RM Miranda, AHL Lima and JLSGV Junior. Using accelerated molecular dynamics simulation to elucidate the effects of the T198F mutation on the molecular flexibility of the West Nile virus envelope protein. Sci. Rep. 2020; 10, 9625.
M Batool, M Shah, MC Patra, D Yesudhas and S Choi. Structural insights into the Middle East respiratory syndrome coronavirus 4a protein and its dsRNA binding mechanism. Sci. Rep. 2017; 7, 11362.
U Doshi, MJ Holliday, EZ Eisenmesser and D Hamelberg. Dynamical network of residue-residue contacts reveals coupled allosteric effects in recognition, catalysis, and mutation. Proc. Natl. Acad. Sci. 2016; 113, 4735-40.
K Kasahara, I Fukuda and H Nakamura. A novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 Dimer-DNA complex. PLoS One 2014; 9, e112419.
D Ni, K Song, J Zhang and S Lu. Molecular dynamics simulations and dynamic network analysis reveal the allosteric unbinding of monobody to H-Ras triggered by R135K mutation. Int. J. Mol. Sci. 2017; 18, 2249.
Downloads
Published
Issue
Section
License
Copyright (c) 2022 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.






