Structural Properties of Monoamine Oxidase B Complexed with Adrenaline in the Deamination Reaction: A Theoretical Study
DOI:
https://doi.org/10.48048/tis.2022.4329Keywords:
Monoamine Oxidase B, Adrenaline, ONIOM Technique, Deamination Reaction, Hydride Transfer, Nucleophilic Mechanism, InhibitorAbstract
Monoamine Oxidase B (MAO B) is a flavoenzyme that regulates and oxidizes monoamine neurotransmitters such as adrenaline, noradrenaline and dopamine. The oxidation reaction also produces hydrogen peroxide that can be harmful to dopaminergic neurons leading to neurological disorders such as Alzheimer’s disease and Parkinson’s disease. MAO B inhibitor is consequently used for preventing those neurological disorders. The structural details insight into the oxidation reaction mechanism of MAO B are vital to design specific inhibitors. A theoretical study is used to explain structural details of theoretical states in molecular level. In this work, the deamination reaction of adrenaline by MAO B was carried out to gain molecular-level details via ONIOM technique at M062X/6–31+G(d,p):PM6 level of theory. According to ONIOM calculation results, there are 4 key MAO B active site residues which are Leu171, Phe343, Tyr398 and Tyr435. Residues that bind adrenaline via hydrophobic interactions are Leu171, Phe343 and Tyr398 while Tyr435 binds via hydrogen bond. Additionally, NBO analysis suggests that the mechanism of this reaction is a hybrid between hydride transfer and polar nucleophilic characters.
HIGHLIGHTS
- The structural properties along the adrenaline deamination reaction by MAO B have been revealed by ONIOM technique
- Key MAO B active site residues for designing more specific MAO B inhibitors have been identified in this study
- The mechanism of adrenaline deamination reaction by MAO B is a mixture between hydride transfer mechanism and polar nucleophilic mechanism
GRAPHICAL ABSTRACT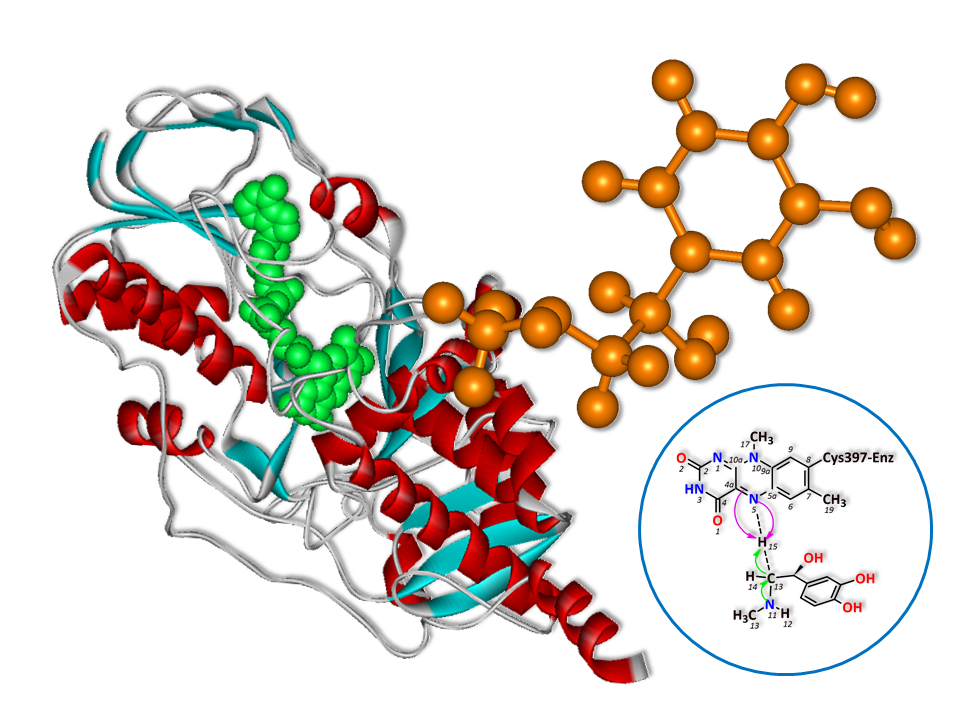
Downloads
References
C Binda, P Newton-Vinson, F Hubálek, DE Edmondson and A Mattevi. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Struct. Biol. 2002; 9, 22-6.
H Gaweska and PF Fitzpatrick. Structures and mechanism of the monoamine oxidase family. Biomol. Concepts 2011; 2, 365-77.
JS Fowler, ND Volkow, GJ Wang, J Logan, N Pappas, C Shea and R MacGregor. Age-related increases in brain monoamine oxidase B in living healthy human subjects. Neurobiol. Aging 1997; 18, 431-5.
K Cakir, SS Erdem and VE Atalay. ONIOM calculations on serotonin degradation by monoamine oxidase B: Insight into the oxidation mechanism and covalent reversible inhibition. Org. Biomol. Chem. 2016; 14, 9239-52.
LW Chung, WMC Sameera, R Ramozzi, AJ Page, M Hatanaka, GP Petrova, TV Harris, X Li, Z Ke, F Liu, H Li, L Ding and K Morokuma. The ONIOM method and its applications. Chem. Rev. 2015; 115, 5678-796.
C Binda, F Hubálek, M Li, DE Edmondson and A Mattevi. Crystal structure of human monoamine oxidase b, a drug target enzyme monotopically inserted into the mitochondrial outer membrane. FEBS Lett. 2004; 564, 225-8.
AE Reed, LA Curtiss and F Weinhold. Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem. Rev. 1988; 88, 899-926.
MJ Frisch, GW Trucks, HB Schlegel, GE Scuseria, MA Robb, JR Cheeseman, G Scalmani, V Barone, GA Petersson, H Nakatsuji, X Li, M Caricato, A Marenich, J Bloino, BG Janesko, R Gomperts, B Mennucci, HP Hratchian, JV Ortiz, AF Izmaylov, …, DJ Fox. Gaussian 09. Gaussian, Inc., Wallingford, 2016.
S Salentin, S Schreiber, VJ Haupt, MF Adasme and M Schroeder. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015; 43, W443-W447.
R Fährrolfes, S Bietz, F Flachsenberg, A Meyder, E Nittinger, T Otto, A Volkamer and M Rarey. ProteinsPlus: A web portal for structure analysis of macromolecules. Nucleic Acids Res. 2017; 45, W337-W343.
M Li, C Binda, A Mattevi and DE Edmondson. Functional role of the “aromatic cage” in human monoamine oxidase B: Structures and catalytic properties of Tyr435 mutant proteins. Biochemistry 2006; 45, 4775-84.
EC Ralph, JS Hirschi, MA Anderson, WW Cleland, DA Singleton and PF Fitzpatrick. Insights into the mechanism of favoprotein-catalyzed amine oxidation from nitrogen isotope effects on the reaction of N-methyltryptophan oxidase. Biochemistry 2007; 46, 7655-64.
CWM Kay, HE Mkami, G Molla, L Pollegioni and RR Ramsay. Characterization of the covalently bound anionic flavin radical in monoamine oxidase a by electron paramagnetic resonance. J. Am. Chem. Soc. 2007; 129, 16091-7.
SS Erdem, Ö Karahan, İ Yıldız and K Yelekçi. A computational study on the amine-oxidation mechanism of monoamine oxidase: Insight into the polar nucleophilic mechanism. Org. Biomol. Chem. 2006; 4, 646-58.
JR Miller and DE Edmondson. Structure-activity relationships in the oxidation of para-substituted benzylamine analogues by recombinant human liver monoamine oxidase A. Biochemistry 1999; 38, 13670-83.
Downloads
Published
Issue
Section
License
Copyright (c) 2022 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.