Insights into Binding Affinity of Flavonoid Compounds from Thai Herbs against 2009 H1N1 Hemagglutinin
DOI:
https://doi.org/10.48048/tis.2022.2689Keywords:
Hemagglutinin, Epicatechin gallate, Puerarin, Docking, MD simulationAbstract
Outbreak of influenza virus is one of serious concerns for public health. Hemagglutinin (HA), a spike-shaped glycoprotein on the viral surface, plays an important role during the early stage of influenza infection. In the present work, a set of flavonoids were screened against 2009 H1N1 HA by computational chemistry techniques. Among 35 flavonoids, the docking results showed that the epicatechin gallate (ECG) and puerarin exhibited a good binding affinity towards 2009 H1N1 HA. These 2 compounds were then studied by all-atom molecular dynamics (MD) simulations. The predicted binding free energy of the H1-puerarin complex (–25.86 ± 2.92 kcal/mol) was slightly greater than that of H1-ECG (–22.81 ± 2.19 kcal/mol), suggesting that the puerarin and ECG could provide similar binding affinity towards 2009 H1 HA target. However, the stronger electrostatic energy contribution of ~10 kcal/mol was found in the puerarin binding to 2009 H1N1 HA. This molecular information of ligand-protein interaction could be helpful in further drug design and development for influenza treatment.
HIGHLIGHTS
- The 35 flavonoid bioactive compounds were screened against 2009 H1N1 HA by molecular docking
- The epicatechin gallate (ECG) and puerarin exhibited a good binding affinity and were then studied by all-atom molecular dynamics (MD) simulations
- The predicted binding free energy revealed that the puerarin and ECG could provide similar binding affinity towards 2009 H1 HA target
GRAPHICAL ABSTRACT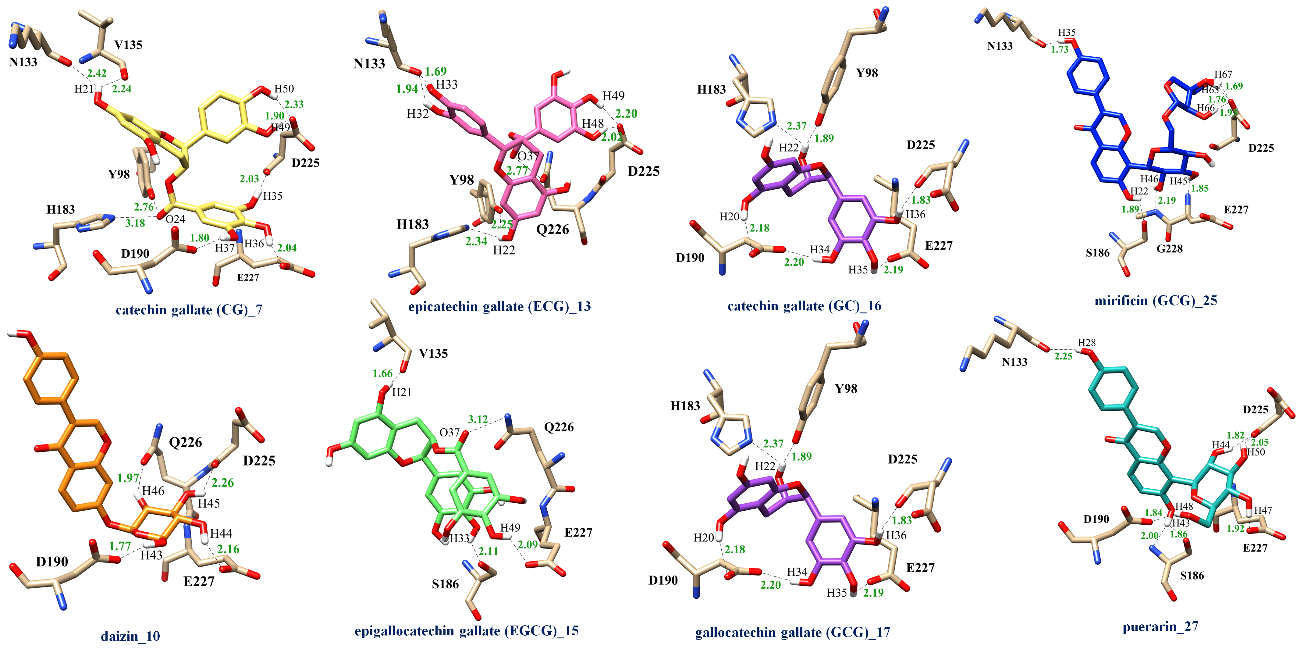
Downloads
References
JJ Skehel and DC Wiley. Influenza viruses and cell membranes. Am. J. Respir. Crit. Care Med. 1995; 152, S13-S15.
T Lin, G Wang, A Li, Q Zhang, C Wu, R Zhang, Q Cai, W Song and KY Yuen. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology 2009; 392, 73-81.
DA Steinhauer. INFLUENZA pathways to human adaptation. Nature 2013; 499, 412-3.
DA Steinhauer. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999; 258, 1-20.
Y Su, L Meng, J Sun, W Li, L Shao, K Chen, D Zhou, F Yang and F Yu. Design, synthesis of oleanolic acid-saccharide conjugates using click chemistry methodology and study of their anti-influenza activity. Eur. J. Med. Chem. 2019; 182, 111622.
Z Jin, Y Wang, XF Yu, QQ Tan, SS Liang, T Li, H Zhang, PC Shaw, J Wang and C Hu. Structure-based virtual screening of influenza virus RNA polymerase inhibitors from natural compounds: Molecular dynamics simulation and MM-GBSA calculation. Comput. Biol. Chem. 2020; 85, 107241.
H Li, M Li, R Xu, S Wang, Y Zhang, L Zhang, D Zhou and S Xiao. Synthesis, structure activity relationship and in vitro anti-influenza virus activity of novel polyphenol-pentacyclic triterpene conjugates. Eur. J. Med. Chem. 2019; 163, 560-68.
S Onishi, T Mori, H Kanbara, T Habe, N Ota, Y Kurebayashi and T Suzuki. Green tea catechins adsorbed on the murine pharyngeal mucosa reduce influenza A virus infection. J. Funct. Foods. 2020; 68, 103894.
C Seniya, S Shrivastava, SK Singh, GJ Khan. Analyzing the interaction of a herbal compound Andrographolide from Andrographis paniculata as a folklore against swine flu (H1N1). Asian Pacific J. Trop. Dis. 2014; 4, S624-S630.
R Latif and CY Wang. Andrographolide as a potent and promising antiviral agent. Chin. J. Nat. Med. 2020; 18, 760-9.
WC Hsu, SP Chang, LC Lin, CL Li, CD Richardson, CC Lin and LT Lin. Limonium sinense and gallic acid suppress hepatitis C virus infection by blocking early viral entry. Antiviral Res. 2015; 118, 139-47.
BS Hwang, IK Lee, HJ Choi and BS Yun. Anti-influenza activities of polyphenols from the medicinal mushroom Phellinus baumii. Bioorg. Med. Chem. Lett. 2015; 25, 3256-60.
R Fioravanti, I Celestino, R Costi, GC Crucitti, L Pescatori, L Mattiello, E Novellino, P Checconi, AT Palamara, L Nencioni and RD Santo. Effects of polyphenol compounds on influenza A virus replication and definition of their mechanism of action. Bioorgan. Med. Chem. 2012; 20, 5046-52.
S Kannan and P Kolandaivel. Antiviral potential of natural compounds against influenza virus hemagglutinin. Comput. Biol. Chem. 2017; 71, 207-18.
L Meng, Y Su, F Yang, S Xiao, Z Yin, J Liu, J Zhong, D Zhou and F Yu. Design, synthesis and biological evaluation of amino acids-oleanolic acid conjugates as influenza virus inhibitors. Bioorgan. Med. Chem. 2019; 27, 115147.
YT Wang, CH Chan, ZY Su and CL Chen. Homology modeling, docking, and molecular dynamics reveal HR1039 as a potent inhibitor of 2009 A(H1N1) influenza neuraminidase. Biophy. Chem. 2010; 147, 74-80.
P Kongsune, S Hannongbua. The role of conserved QXG and binding affinity of S23G & S26G receptors on avian H5, swine H1 and human H1 of influenza A virus hemagglutinin. J. Mol. Graph. Model. 2018; 82, 12-19.
FC Bernstein, TF Koetzle, GJ Williams, EFJ Meyer, MD Brice, JR Rodgers, O Kennard, T Shimanouchi, M Tasumi. The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 1977; 112, 535-42.
R Xu, R McBride, CM Nycholat, JC Paulson, IA Wilson. Structural Characterization of the Hemagglutinin Receptor Specificity from the 2009 H1N1 Influenza Pandemic. J. Virol. 2012; 86, 982-90.
R Dennington, T Keith and J Millam. GaussView. Semichem Inc., Shawnee Mission KS., 2009.
MJ Frisch, M Frisch, G Trucks, K. Schlegel, G Scuseria, M Robb, J Cheeseman, J Montgomery, T Vreven, KN Kudin, J Burant, J Millam, S Iyengar, J Tomasi, V Barone, B Mennucci, M Cossi, G Scalmani, N Rega, A Petersson, H Nakatsuji, M Hada, M Ehara, K Toyota, R Fukuda, J Hasegawa, M Ishida, T Nakajima, Y Honda, O Kitao, H Nakai, M Klene, X Li, J Knox, H Hratchian, D Cross, V Bakken, C Adamo, J Jaramillo-Merchan, R Gomperts, R Stratmann, O Yazyev, A Austin, R Cammi, C Pomelli, J Ochterski, P Ayala, K Morokuma, G Voth, P Salvador, JJ Dannenberg, VG Zakrzewski, S Dapprich, AD Daniels, M Strain, O Farkas, S Malick, A Rabuck, K Raghavachari, J Foresman, J Ortiz, Q Cui, AG Baboul, S Clifford, J Cioslowski, B Stefanov, G Liu, A Liashenko, P Piskorz, I Komaromi, R Mata, D Fox, T Keith, S Laham, CY Peng, A Nanayakkara, M Challacombe, P Gill, B Johnson, W Chen, M Wong, RS González, J Pople, J Dannenberg, V Zakrzewski, A Daniels, AG Baboul, Y Peng, GE Scuseria, JM Millam, JB Foresman, MJ Frisch, JA Montgomery, RE Stratmann, DK Malick, XB Li, T Keith, MW. Wong, HB Schlegel, PY Ayala, BB Stefanov, M Hada, RL Martin, KN Kudin, HP Hratchian, GA Voth, AJ Austin, MC Strain, C Adamo, PMW Gill, MA Robb, GA Petersson, JB Cross, JL Torre, GW Trucks, JC Burant, DJ Fox,bAD Rabuck, C Huerta, M Akhras, JR Cheeseman, SS Iyengar, JA Pople, JE Knox, JW Ochterski and BA Johnson. Gaussian 03, revision C.02, Gaussian, Inc., Wallingford CT, 2004.
GM Morris, DS Goodsell, RS Halliday, R Huey, WE Hart, RK Belew and AJ Olson. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 1998; 19, 1639-62.
J Gasteiger and M Marsili. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron 1980; 36, 3219-28.
GM Morris, R Huey, W Lindstrom, MF Sanner, RK Belew, DS Goodsell and AJ Olson. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009; 30, 2785-91.
N Nunthaboot, T Rungrotmongkol, M Malaisree, N Kaiyawet, P Decha, P Sompornpisut, Y Poovorawan and S Hannongbua. Evolution of human receptor binding affinity of H1N1 hemagglutinins from 1918 to 2009 pandemic influenza A virus. J. Chem. Inf. Model. 2010; 50, 1410-17.
R Huey, GM Morris, AJ Olson and DS Goodsel. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007; 28, 1145-52.
E Ermakova. Structural insight into the glucokinase-ligands interactions: Molecular docking study. Comput. Biol. Chem. 2016; 64, 281-96.
D Case, R Betz, DS Cerutti, T Cheatham, T Darden, R Duke, TJ Giese, H Gohlke, A Götz, N Homeyer, S Izadi, P Janowski, J Kaus, A Kovalenko, TS Lee, S LeGrand, P Li, C Lin, T Luchko and P Kollman. Amber 16. University of California, San Francisco, 2016.
WD Cornell, P Cieplak, CI Bayly and PA Kollmann. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 1993; 115, 9620-31.
Y Duan, C Wu, S Chowdhury, MC Lee, G Xiong, W Zhang, R Yang, P Cieplak, R Luo, T Lee, J Caldwell, J Wang and P Kollman. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003; 24, 1999-2012.
WD Cornell, P Cieplak, CI Bayly, IR Gould, KM Merz, DM Ferguson, DC Spellmeyer, T Fox, JW Caldwell and PA Kollman. A second generation forcefield for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995; 117, 5179-97.
JM Wang, W Wang and PA Kollman. Antechamber: An accessory software package for molecular mechanical calculations. J. Am. Chem. Soc. 2001; 222, U403-U403.
DC Bas, DM Rogers and JH Jensen. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins 2008; 73, 765-83.
H Li, AD Robertson and JH Jensen. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005; 61, 704-21.
HJC Berendsen, JPM Postma, WF Van Gunsteren, A DiNola and JR Haak. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984; 81, 3684-90.
JP Ryckaert, G Cicotti and HJC Berendsen. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977; 23, 327-41.
DM York, TA Darden and LG Pedersen. The effect of long-range electrostatic interactions in simulations of macromolecular crystals: a comparison of the Ewald and truncated list methods. J. Chem. Phys. 1993; 99, 8345-8.
DR Roe and TE Cheatham. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory. Comput. 2013; 9, 3084-95.
N Nunthaboot, T Rungrotmongkol, M Malaisree, P Decha, N Kaiyawet, P Intharathep, P Sompornpisut, Y Poovorawan, S Hannongbua. Molecular insights into human receptor binding to 2009 H1N1 influenza A hemagglutinin. Monatsh Chem. 2009; 141, 801-7.
HX Wang, MS Zeng, Y, Ye, JY Liu and PP Xu. Antiviral activity of puerarin as potent inhibitor of influenza virus neuraminidase. Phytother. Resh. 2021; 35, 324-36.
Downloads
Published
Issue
Section
License

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.