Structure-Based Design and Molecular Simulations of Some Quercetin-Based Drugs as Repurposable Inhibitors of SARS-CoV-2 Main Protease
DOI:
https://doi.org/10.48048/tis.2022.2557Keywords:
SARS-CoV-2 main protease, Molecular docking, Molecular dynamics, Quercetin, Natural repurposable drugsAbstract
In this study, virtual screening and molecular dynamics (MD) protocols were applied to screen 2826 FDA-approved natural product drugs from the Selleckchem.com library for prospective inhibitors of the SARS-CoV-2 main protease. From the virtual screening through HTVS, SP and XP docking analysis, hyperoside, rutin hydrate, rutoside and quercitrin displayed a stronger binding affinity with respective XP docking scores of –11.389, –11.340, –11.087 and –10.232 kcal/mol than co-crystallized N-[2-(5-fluoro-1H-indol-3-yl)ethyl]acetamide (HWH) and positive inhibitors, lopinavir and ritonavir which scored –5.493, –6.463 and –6.221 kcal/mol respectively. Selectively, the binding free energy, MMGB(SA) of hyperoside and rutin hydrate was observed as –21.55 and –25.82 kcal/mol respectively compared to lopinavir and ritonavir with –17.66 and –5.28 kcal/mol respectively. Consistently, the selected drugs displayed good thermodynamics conformational stability, thus, recommended as promising repurposable inhibitors of the SARS-CoV-2 main protease amenable for further studies.
HIGHLIGHTS
- Main protease of the SARS-CoV-2 represents a plausible target for mitigating viral replication
- Natural repurposable drugs are safer and cheaper for combating COVID-19
- Hyperoside, rutin hydrate and quercitrin displayed strong binding and stability with the viral main protease
- They are recommended as repurposable inhibitors of SARS-CoV-2 main protease for further studies
GRAPHICAL ABSTRACT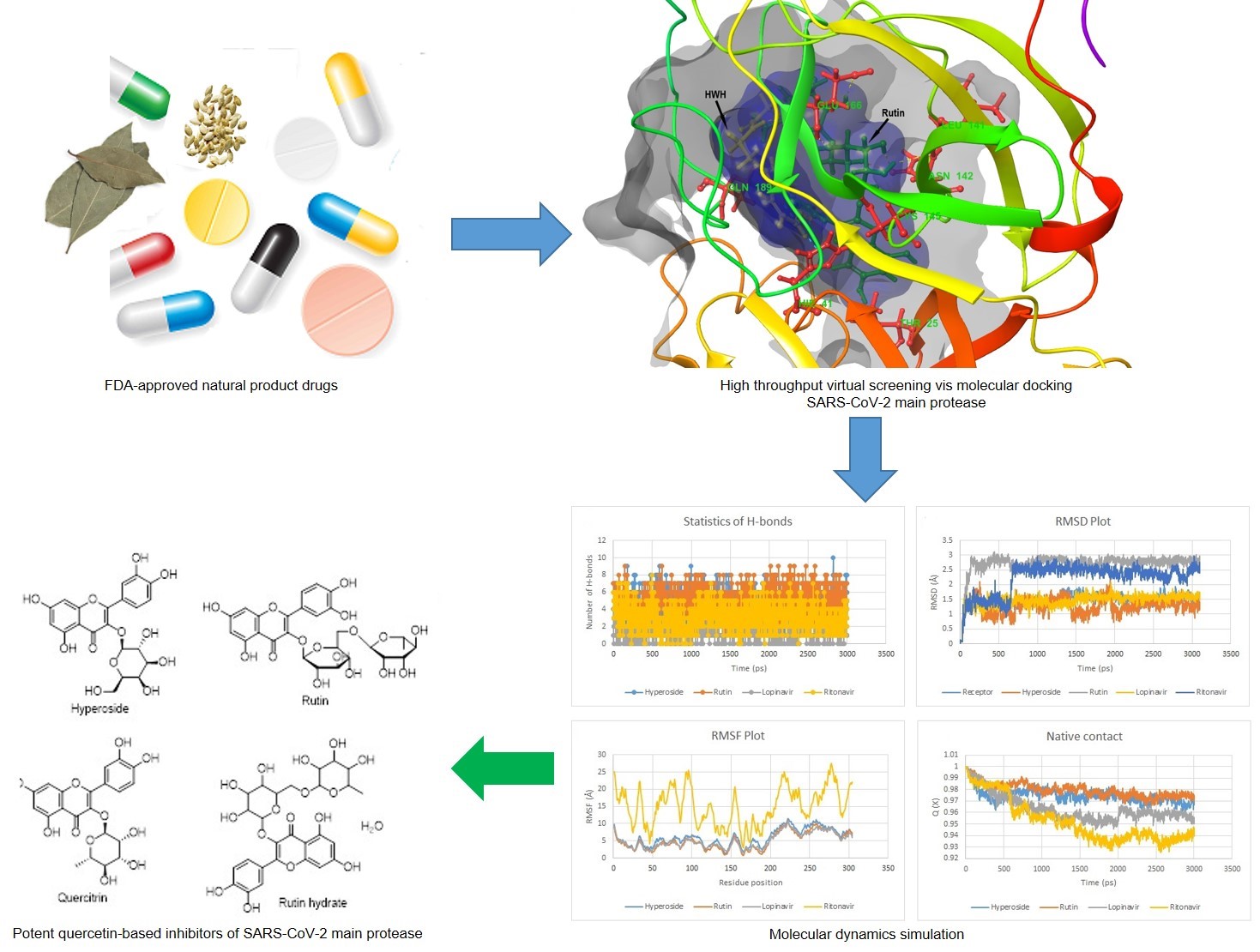
Downloads
References
K Amporndanai, X Meng, W Shang, Z Jin, M Rogers, Y Zhao, Z Rao, ZJ Liu, H Yang, L Zhang, PM O'Neill and SS Hasnain. Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat. Commun. 2021; 12, 3061.
A Douangamath, D Fearon, P Gehrtz, T Krojer, P Lukacik, CD Owen, E Resnick, C Strain-Damerell, A Aimon, P Ábrányi-Balogh, J Brandão-Neto, A Carbery, G Davison, A Dias, TD Downes, L Dunnett, M Fairhead, JD Firth, SP Jones, … MA Walsh. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020; 11, 5047.
YO Ayipo, SN Yahaya, WA Alananzeh, HF Babamale and MN Mordi. Pathomechanisms, therapeutic targets and potent inhibitors of some beta-coronaviruses from bench-to-bedside. Infect. Genet. Evol. 2021; 93, 104944.
YO Ayipo, I Ahmad, YS Najib, SK Sheu, H Patel and MN Mordi. Molecular modelling and structure-activity relationship of a natural derivative of o-hydroxybenzoate as a potent inhibitor of dual NSP3 and NSP12 of SARS-CoV-2: in silico study. J. Biomol. Struct. Dyn. 2022. DOI: 10.1080/07391102. 2022.2026818.
Recovery Collaborative Group. Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2020; 396, 1345-52.
V Asati, SS Thakur, N Upmanyu and SK Bharti. Virtual screening, molecular docking, and DFT studies of some thiazolidine-2,4-diones as potential PIM-1 kinase inhibitors. ChemistrySelect 2018; 3, 127-35.
DL Lynch, DP Hurst, DM Shore, MC Pitman and PH Reggio. Molecular dynamics methodologies for probing cannabinoid ligand/receptor interaction. Methods Enzymol. 2017; 593, 449-90.
AAAMM Alkhatip, M Georgakis, LRM Valenzuela, M Hamza, E Farag, J Hodgkinson, H Hosny, AM Kamal, M Wagih, A Naguib, H Yassin, H Algameel, M Elayashy, M Abdelhaq, MI Younis, H Mohamed, M Abdulshafi and MA Elramely. Metal-bound methisazone; novel drugs targeting prophylaxis and treatment of sars-cov-2, a molecular docking study. Int. J. Mol. Sci. 2021; 22, 2977.
RA Friesner, JL Banks, RB Murphy, TA Halgren, JJ Klicic, DT Mainz, MP Repasky, EH Knoll, M Shelley, JK Perry, DE Shaw, P Francis and PS Shenkin. Glide: A new approach for rapid, accurate docking and scoring. 1. method and assessment of docking accuracy. J. Med. Chem. 2004; 47, 1739-49.
TA Halgren, RB Murphy, RA Friesner, HS Beard, LL Frye, WT Pollard and JL Banks. Glide: A new approach for rapid, accurate docking and scoring. 2. enrichment factors in database screening. J. Med. Chem. 2004; 47, 1750-9.
W Cui, K Yang and H Yang. Recent progress in the drug development targeting SARS-CoV-2 main protease as treatment for COVID-19. Front. Mol. Biosci. 2020; 7, 616341.
Y Kumar, H Singh and CN Patel. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation-based drug-repurposing. J. Infect. Public Health 2020; 13, 1210-23.
E Harder, W Damm, J Maple, C Wu, M Reboul, JY Xiang, L Wang, D Lupyan, MK Dahlgren, JL Knight, JW Kaus, DS Cerutti, G Krilov, WL Jorgensen, R Abel and RA Friesner. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016; 12, 281-96.
GM Sastry, M Adzhigirey, T Day, R Annabhimoju and W Sherman. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013; 27, 221-34.
SV Stoddard, SD Stoddard, BK Oelkers, K Fitts, K Whalum, K Whalum, AD Hemphill, J Manikonda, LM Martinez, EG Riley, CM Roof, N Sarwar, DM Thomas, E Ulmer, FE Wallace, P Pandey and S Roy. Optimization rules for SARS-CoV-2 Mpro antivirals: Ensemble docking and exploration of the coronavirus protease active site. Viruses 2020; 12, 942.
M Luo, H Wang, Y Zou, S Zhang, J Xiao, G Jiang, Y Zhang and Y Lai. Identification of phenoxy acetamide derivatives as novel DOT1L inhibitors via docking screening and molecular dynamics simulation. J. Mol. Graph. Model. 2016; 68, 128-39.
GK Veeramachaneni, KK Raj, LM Chalasani, JS Bondili and VR Talluri. High-throughput virtual screening with e-pharmacophore and molecular simulations study in the designing of pancreatic lipase inhibitors. Drug Des. Devel. Ther. 2015; 9, 4397-412.
JF Yang, F Wang, YZ Chen, GF Hao and GF Yang. LARMD: Integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of the estrogen receptor. Brief Bioinform. 2019; 21, 2206-18.
YO Ayipo, SN Yahaya, HF Babamale, I Ahmad, H Patel and MN Mordi. β-Carboline alkaloids induce structural plasticity and inhibition of SARS-CoV-2 nsp3 macrodomain more potently than remdesivir metabolite GS-441524: Computational approach. Turk. J. Biol. 2021; 45, 503-17.
Z Babar, M Khan, M Zahra, M Anwar, K Noor, HF Hashmi, M Suleman, M Waseem, A Shah, S Ali and SS Ali. Drug similarity and structure-based screening of medicinal compounds to target macrodomain- I from SARS-CoV-2 to rescue the host immune system: A molecular dynamics study. J. Biomol. Struct. Dyn. 2020; 40, 523-37.
S Genheden and U Ryde. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015; 10, 449-61.
E Wang, H Sun, J Wang, Z Wang, H Liu, JZH Zhang and T Hou. End-point binding free energy calculation with MM/PBSA and MM/GBSA: Strategies and applications in drug design. Chem. Rev. 2019; 119, 9478-508.
J Zhao, Y Cao and L Zhang. Exploring the computational methods for protein-ligand binding site prediction. Comput. Struct. Biotechnol. J. 2020; 18, 417-26.
A Castro-Alvarez, AM Costa and J Vilarrasa. The performance of several docking programs at reproducing protein-macrolide-like crystal structures. Molecules 2017; 22, 136.
D Ramírez and J Caballero. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules 2018; 23, 1038.
P Parmar, P Rao, A Sharma, A Shukla, RM Rawal, M Saraf, BV Patel and D Goswami. Meticulous assessment of natural compounds from NPASS database for identifying analogue of GRL0617, the only known inhibitor for SARS‑CoV2 papain-like protease (PLpro) using rigorous computational workflow. Mol. Divers. 2022; 26, 389-407.
CS Silva, GV Portari and H Vannucchi. Antioxidant treatment and alcoholism. In: VB Patel (Ed.). Molecular aspects of alcohol and nutrition. Academic Press, London, 2016, p. 119-31.
JK Aronson. Iodinated contrast media. In: Meyler's side effects of drugs. 16th eds. Elsevier Science, Amsterdam, The Netherlands, 2016, p. 239-97.
JM Terrell and TF Jacobs. Alogliptin. In: StatPearls: Content is king. StatPearls Publishing, Treasure Island, Florida, 2021, Available at: https://www.ncbi.nlm.nih.gov/books/NBK507809, accessed January 2022.
T Dai, J Chen, DJ McClements, P Hu, X Ye, C Liu and T Li. Protein-polyphenol interactions enhance the antioxidant capacity of phenolics: Analysis of rice glutelin-procyanidin dimer interactions. Food Funct. 2019; 10, 765-74.
L Seczyk, M Swieca, I Kapusta and U Gawlik-Dziki. Protein-phenolic interactions as a factor affecting the physicochemical properties of white bean proteins. Molecules 2019; 24, 408.
U.S. National Library of Medicine. Clinical trials on lopinavir and ritonavir for COVID-19. 2021, Available at: https://www.clinicaltrials.gov/ct2/results?cond=Covid19&term=lopinavir&cntry=& state=&city=&dist =, accessed June 2021.
G Klebe. Protein-ligand interactions as the basis for drug action. In: G Klebe (Ed.). Drug design. Springer, Berlin, Heidenberg, Germany, 2013, p. 61-88.
AS Panja, S Maiti and B Bandyopadhyay. Protein stability governed by its structural plasticity is inferred by physicochemical factors and salt bridges. Sci. Rep. 2020; 10, 1822.
A Saeedi-boroujeni and MR Mahmoudian-Sani. Anti-inflammatory potential of Quercetin in COVID-19 treatment. J. Inflamm. 2021; 18, 3.
R Shukla, V Pandey, GP Vadnere and S Lodhi. Role of flavonoids in management of inflammatory disorders. In: VR Preedy (Ed.). Bioactive food as dietary interventions for arthritis and related inflammatory diseases. 2nd eds. Academic Press, London, 2019, p. 293-322.
RJ Horwitz. The allergic patient. In: Integrative medicine. 4th eds. Elsevier, Amsterdam, The Netherlands, 2018, p. 300-9.
Downloads
Published
Issue
Section
License
Copyright (c) 2022 Walailak University

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.